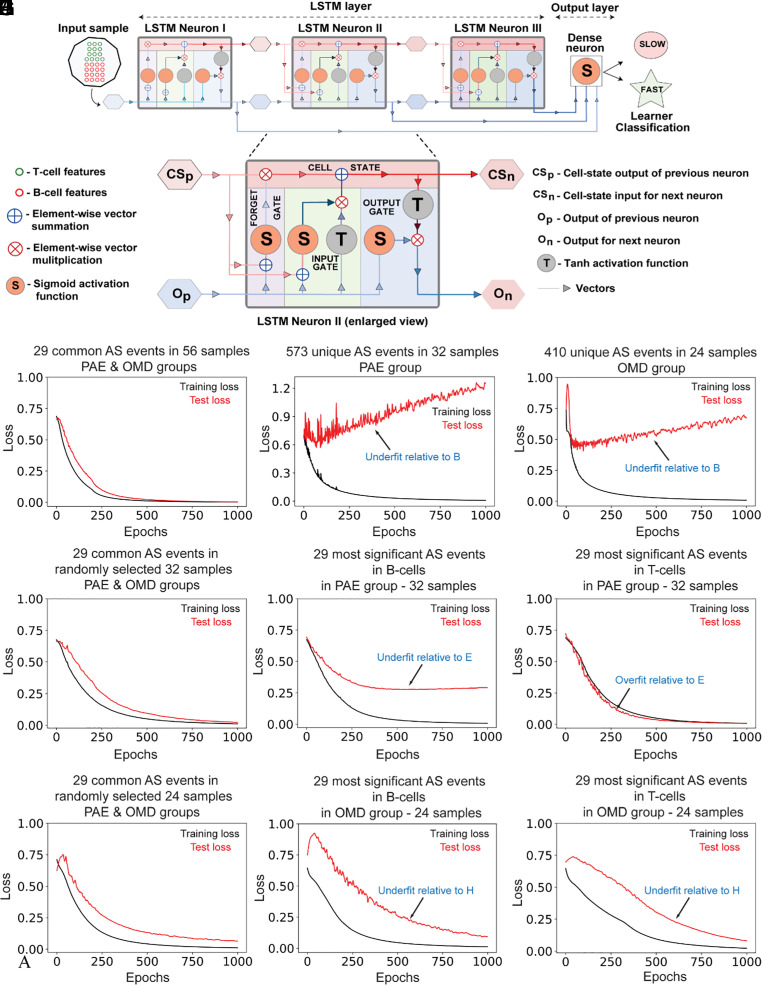
Fig. 4.
Psi values of AS events common to PAE and OMD are better than those unique to PAE and OMD at adequately training a deep-learning model to accurately predict motor learner type. (A) LSTM model architecture consisting of an LSTM layer with three LSTM neurons and an output layer with one dense neuron. Each LSTM neuron comprises four gates: an input gate for information input, a forget gate for forgetting irrelevant learned associations, a cell-state (memory) gate for remembering relevant learned associations, and an output gate for output of learned information. Vectors show flow of information and their various permutations during learning. For training and testing the LSTM model, psi values of 29 common AS events were used as input, and learnability of PAE and OMD mice, classified as either slow or fast learners (learning-index score below or above the population median of 2.8, respectively), was used as output. Of note, 80% and 20% of input was used for training and testing respectively. (B–J) Comparison of learnability of the LSTM model, as assessed via distribution of binary cross-entropic loss function values over epochs. Psi values from different subsets of data with equivalent sample sizes, i.e., from the common 29 AS biomarkers (B, E, and H), or all unique significant events in B cells and T cells in PAE (C) or OMD (D), or only the top 29 most significant unique AS events in B cells (F and I) or T cells (G and J) in PAE (F and G) and OMD (I and J), were used as input. For equivalent comparison of model performance across B–J, sample number in biomarker group (B, E, and H) was randomly reduced from 56 (B, all samples) to 32 (E) to 24 (H). In all comparisons, AS events in common biomarker group (B, E, and H) resulted in better LSTM model learnability than either all unique significant events (C and D) or the top 29 most unique significant AS events in either PAE (F and G) or OMD (I and J). This worse LSTM model learnability for each comparison is annotated with blue text (C, D, F, G, I, and J) as either underfitting or overfitting relative to LSTM model performance with biomarker group (B, E, and H).