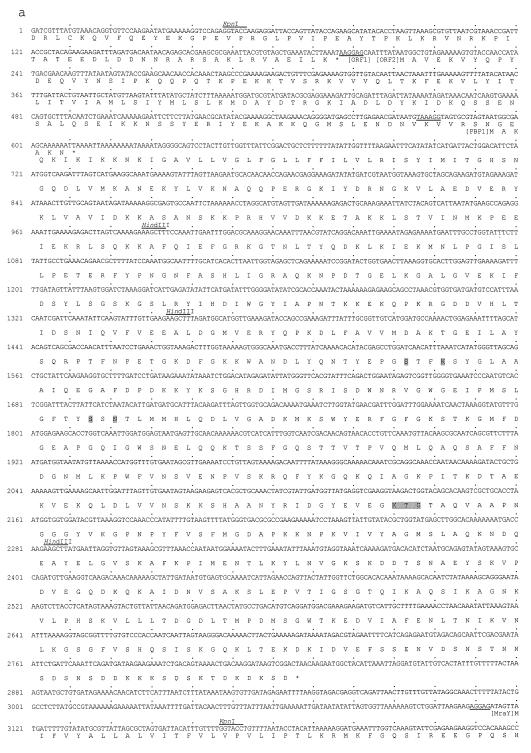
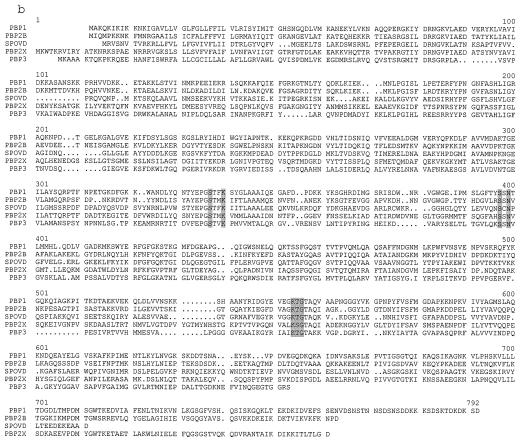
FIG. 3.
(a) Nucleotide sequence of a part of the 4.9-kb fragment of NCTC8325 shown in Fig. 2, and its deduced amino acid sequence. The putative ribosome binding site of each ORF is underlined. The conserved motifs of PBPs are shown by shading in the amino acid sequence of PBP1. KpnI and HindIII sites used for constructing the plasmids shown in Fig. 2 are indicated above the nucleotide sequence. (b) Amino acid sequence alignment of PBPs of various organisms including PBP1 of S. aureus. Common motifs of PBPs are shown by shadowing PBP1, PBP1 of S. aureus; PBP2B, PBP2B of B. subtilis; SPOVD, SpoVD of B. subtilis; PBP2X, PBP2X of S. pneumoniae; PBP3, PBP3 of E. coli.