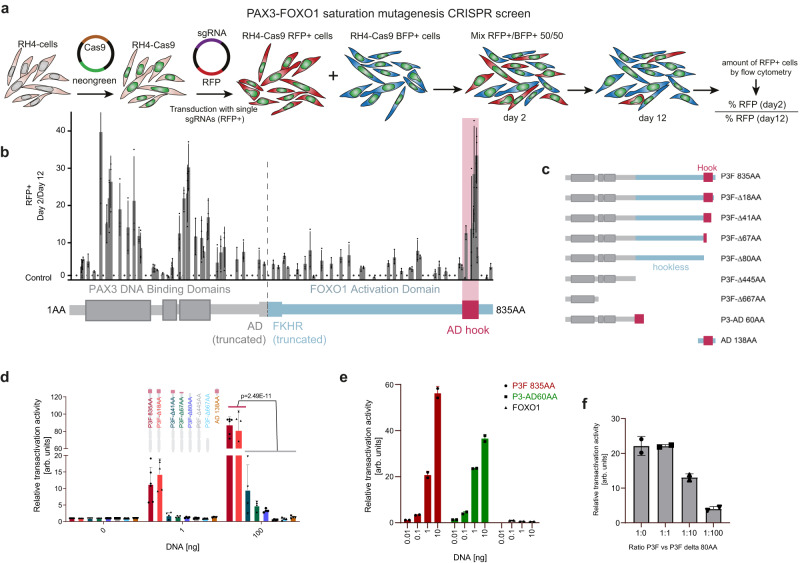
Fig. 1. CRISPR/Cas9-based domain screening of P3F reveals a novel functionally important C-terminal domain.
a Scheme depicting the CRISPR/Cas9-based domain screen approach. RH4 cells stably expressing Cas9 (RH4-Cas9) were transduced with either a vector driving expression of sgRNAs directed against P3F and RFP or a control sgRNA directed against the AAVS1 region together with BFP. Two days after transduction RFP+ cells were mixed 1:1 with BFP+ cells. Percentage of RFP+ cells was determined at day 2 and 12 by flow cytometry. b Ratio of RFP+ cells on day 2 (D2) and day 12 (D12). Relative number of RFP+ cells was measured by flow cytometry. The horizontal dashed line indicates the mean of all controls (ratio 1.22). Plotted are mean and standard deviation for each sgRNA (n = 3 independent experiments). P3F domains (PAX3 in grey, FOXO1 in blue) are depicted schematically at the bottom, with the vertical dashed line indicating the breakpoint of the fusion. AD, activation domain; FKHR, forkhead domain. c Scheme depicting the truncated versions of P3F used for reporter assays. d Luciferase reporter assays measured 48 h after transfection of HEK 293 T cells with indicated P3F constructs. Depicted are mean and standard deviation for each construct normalized to an internal transfection control (Renilla luciferase) (n = 6 (P3F 835 AA), n = 5 (P3F-Δ18AA) and n = 4 (rest) independent experiments; two-way Anova, Tukey’s multiple comparisons test). e Luciferase assay performed as described under d using full-length P3F, a 60 AA fragment containing the AD fused to the PAX3 part of P3F (P3-AD 60AA) or wildtype FOXO1 (n = 2 independent experiments). f Luciferase assay performed as described under d with full-length P3F in combination with the P3F-Δ80AA deletion mutant in indicated ratios (n = 2 independent experiments). Source data are provided as a Source Data file.