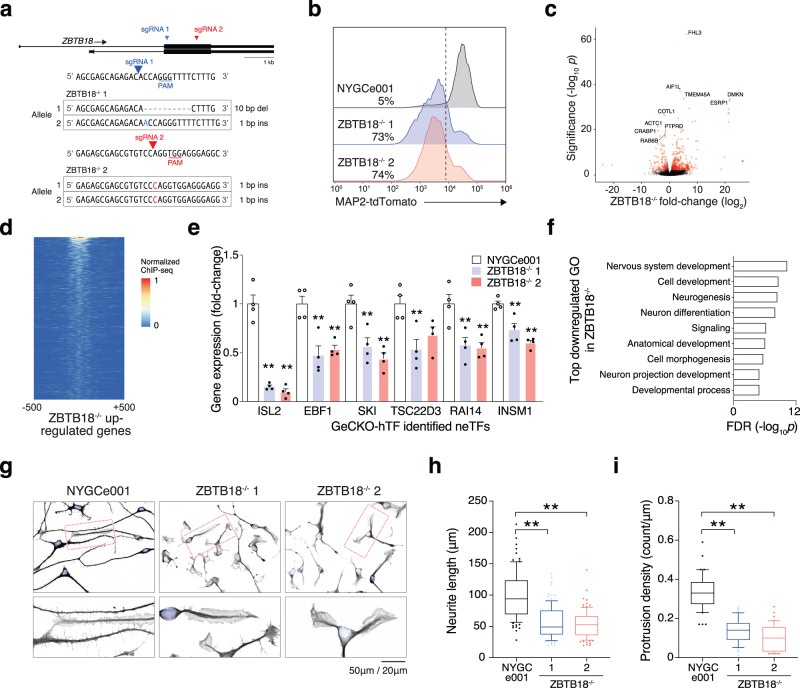
Fig. 5. ZBTB18-null cells have widespread alterations in gene expression and result in immature neurons with stunted neurite development.
a Schematic illustration of the strategy for establishing ZBTB18-null hESCs using CRISPR/Cas9 genome editing with 2 different guide RNAs. b Flow cytometry of MAP2-tdTomato from induced neurons (iNs) at day 7 after NEUROG1/2-induction from NYGCe001 (parental) and ZBTB18-/-−1 and ZBTB18-/-−2 (isogenic knockouts). c Volcano plots for differential gene expression of ZBTB18-/- iNs at day 4 after NEUROG1/2-induction (n = 4 replicates per genotype and timepoint). Red dots indicate genes with padj < 0.01 and |fold-change | > 2. d ZBTB18 binding near upregulated genes via chromatin immunoprecipitation with sequencing in HEK293T cells (n = 2 biological replicates, data is from ref. 32). e Other neuron-essential TFs with significant changes in expression in ZBTB18-/-−1 and ZBTB18-/-−2 at day 4 after NEUROG1/2-induction. f Top-ranked Gene Ontology categories in significantly altered genes in ZBTB18-null iNs at day 4 after NEUROG1/2-induction. g Representative epifluorescence images of WT, ZBTB18-/-−1 and ZBTB18-/-−2 iNs at day 4 after NEUROG1/2-induction. h–i Quantification analysis for neurite length h and protrusion density i for WT, ZBTB18-/-−1 and ZBTB18-/-−2 iNs at day 4 after NEUROG1/2-induction.