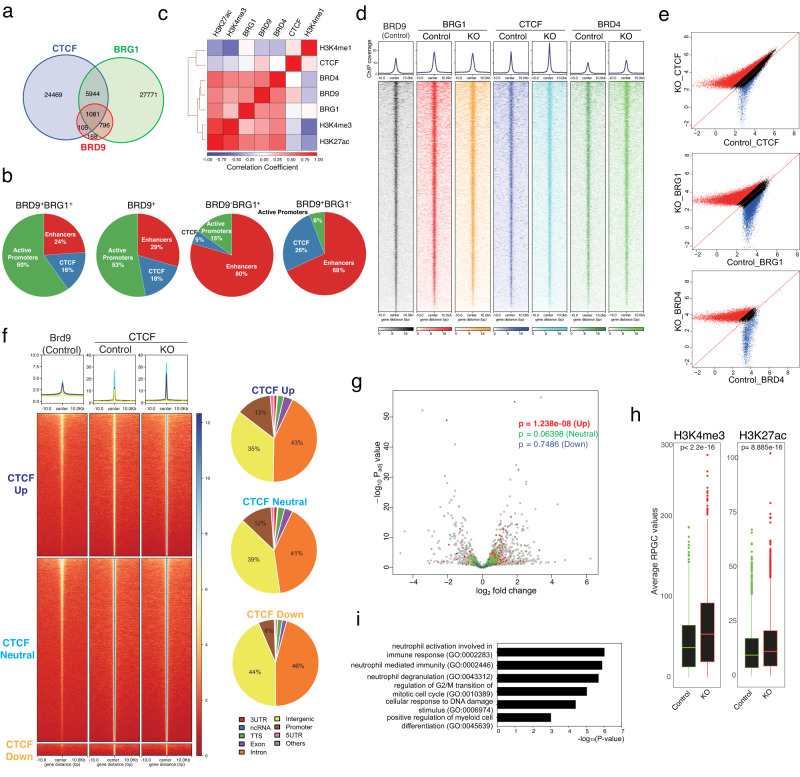
Fig. 5. BRD9 loss enhances CTCF localization resulting in active transcription.
a Venn diagram of peaks from BRD9, BRG1, and CTCF evaluated by ChIP-seq experiments. b The proportion of BRD9+BRG1+(ncBAF), BRD9+ (ncBAF + ncBAF-independent BRD9 targets), BRD9-BRG1+ (BAFs other than ncBAF) and BRD9+BRG1− (ncBAF-independent BRD9 targets) peaks at CTCF, active promoters and enhancers. c Heatmap representing the correlations between average ChIP-seq reads (log2[reads per genome coverage (RPGC)]) over a merged set of BRD9, BRG1, CTCF, BRD4, H3K4me1, H3K4me3, and H3K27ac peaks. d The average ChIP enrichment profile of BRD9, BRG1, CTCF, and BRD4. Heatmap illustrating the ChIP-seq signal 10 kb up and downstream at BRD9 peaks (q < 0.05). e Scatterplot of log2 average RPGC values of CTCF, BRG1, and BRD4 in control vs. KO condition (Red and Blue: ratio of average RPGC values > 1.5, Black: otherwise). f The average ChIP enrichment profile of BRD9 and CTCF (top) over the peak regions in three groups: CTCF Up (dark blue, fold change >1.5), CTCF Neutral (light blue, 1.5 ≥ fold change ≥ 1/1.5), and CTCF Down (yellow, fold change <1/1.5) in KO compared to Control. Heatmap illustrating the ChIP-seq signal 10 kb up and downstream. The genomic distribution of each group is shown on the right. g Volcano plot of DEGs in Brd9 KO HSPCs. Genes whose promoter-TSS sites locate on CTCF peaks in CTCF Up (red), CTCF Neutral (green), CTCF Down (blue) are highlighted and p values of two-sided t-test were indicated. h Correlation between the active histone marks (H3K4me3, left; H3K27ac, right) and the promoter-located CTCF peaks of CTCF Up group in Control and KO conditions. One biologically independent sample in each group was used. p values of two-sided t-test were indicated. In the box-and-whisker plots, the lower and upper hinges correspond to the 25th and 75th percentiles. The whiskers extend from the hinges to the values no further than 1.5 x the inter-quartile ranges. Data points beyond the whiskers are shown as dot plots. i GO terms enrichment of the promoter-located CTCF peaks of CTCF Up group. p values are generated via Enricher. Source data are provided as a Source Data file.