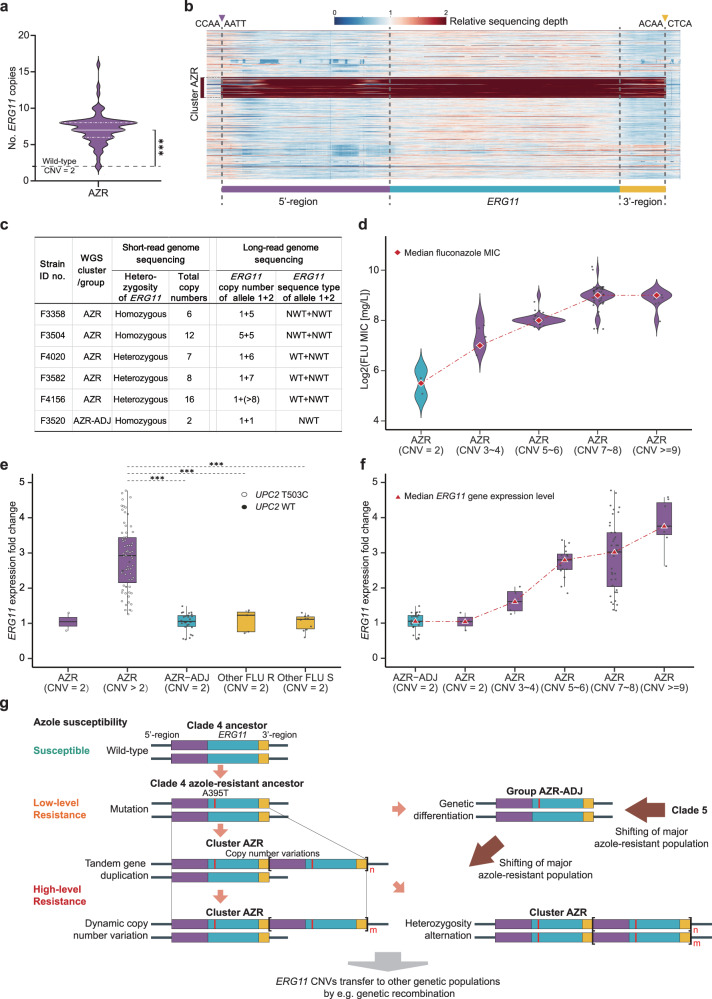
Fig. 7. ERG11 tandem gene duplications observed in C. tropicalis cluster AZR isolates.
a Distribution of ERG11 CNVs observed in 76 cluster AZR isolates. ***p-values < 0.001. b A heatmap of copy number variations (CNVs) of the ERG11 gene observed among 629 isolations by short-read genome sequencing. Each row of the heatmap represents a single strain, and the order of strains from the top to the bottom is in accordance with what appears in Supplementary Data 1. Colors indicate the relative sequencing depth of each base pair within the region shown, using the average sequencing depth of each strain as a reference. The CNV segment identified in cluster AZR isolates comprised the complete 1587 bp ERG11 coding domain sequence (blue), its 1164 bp 5’-upstream region (purple) and 316 bp 3’-downstream region (yellow). Up- and downstream break sequences shared by all strains with CNVs are labeled with purple and yellow triangles at the top. c Long-read sequencing of selected cluster AZR isolates indicated that ERG11 CNVs resulted from tandem gene duplications. Moreover, CNVs occurred only on ERG11 mutant alleles (with the A395T mutation). d Distribution of fluconazole (FLU) MICs to ERG11 CNVs. It was observed that a rise in fluconazole MIC distribution was correlated with an increase in ERG11 CNVs. e Comparison of ERG11 gene expression levels among isolates in different phylogenetic populations. Box-and-whisker plots were used to present gene expression levels, and data points were overlayed as dot plots. The upper and lower whiskers represent maximum and minimum values, respectively; the hinges of the box represent 25% to 75% percentiles and the horizontal line within the box represents median values. Statistical analysis was carried out using two-sided Kruskal-Wallis H test and adjustments were made for multiple comparisons; ***adjusted p-values < 0.001. It was revealed that cluster AZR isolates with ERG11 CNVs had significantly higher ERG11 gene expression levels. f Association between ERG11 gene expression levels and ERG11 CNVs. It demonstrated that an increase in ERG11 CNVs contributed to a significant upregulation of the gene expression. g A proposed evolution history of C. tropicalis clade 4/cluster AZR isolates.