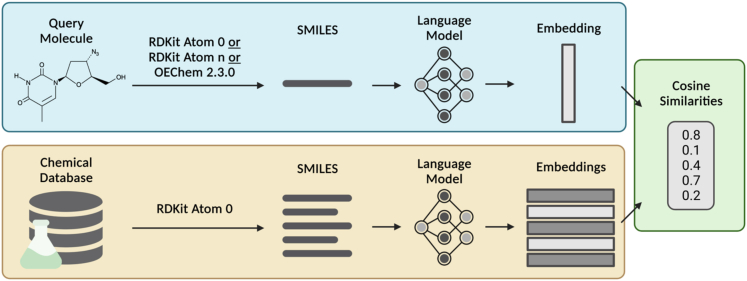
Figure 1.
Chemical semantic search
The query molecule and chemical database are converted into SMILES strings, canonicalized, and then inputted into a language model to obtain embeddings. The cosine similarity between the query embedding and database embeddings is computed, resulting in a vector of embedding similarities. The database is canonicalized with RDKit Atom 0, whereas the queries are canonicalized using one of the following: RDKit Atom 0, RDKit Atom n, or OEChem 2.3.0. Figure created with BioRender.com.