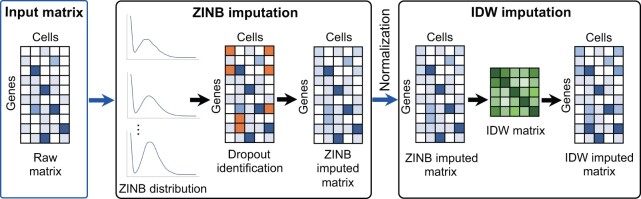
Figure 1.
Flowchart of tsImpute. TsImpute first identifies dropouts by estimating the dropout probability of ZINB distribution, then performs initial ZINB imputation on each dropout by combining dropout probability, expression expectation of the gene, and scale factor of the cell. After initial ZINB imputation, tsImpute divides the cells into different subpopulations using hierarchical clustering and perform IDW imputation for each cluster, which borrows information from cells in the same cluster and weight the information according to the distance between the cells.