Abstract
Objective:
Dendritic cells (DCs) are important players in immunity against pathogens, but overactive DCs have been implicated in autoimmune diseases, like lupus, in which a paucity of targeted therapies remains. Recent research shows that DCs upregulate their immunometabolism when activating. We explored whether modulating fatty acid (FA) metabolism needed for oxidative phosphorylation can affect the activation of two main DC subsets.
Materials & Methods:
Sorted murine plasmacytoid DCs (pDCs) and conventional DCs (cDCs), generated in FLT3-L medium, were treated with etomoxir, an inhibitor of FA oxidation, or TOFA, an inhibitor of FA synthesis, then stimulated with TLR9 agonist CpGA. Surface activation markers and viability were analyzed by flow cytometry, cytokine and chemokine production were measured by ELISA.
Results:
Modulation of FA metabolism suppressed the upregulation of costimulatory molecules and the production of proinflammatory cytokine IL-6 and type I Interferon dependent chemokine CXCL10 by both subsets of DCs, without affecting DC viability, neither of resting DCs or upon activation. Etomoxir inhibited pDCs at lower doses than cDCs, suggesting that pDCs may be more susceptible to FA metabolic modulation.
Conclusions:
Both cDCs, the primary antigen presenting cell, and pDCs, the primary type I IFN producer, exhibit a suppressed ability to activate but normal viability when their FA metabolism is inhibited by etomoxir or TOFA. Our findings indicate that FA metabolism plays an important role in the activation of both pDCs and cDCs and suggest that its modulation is an exploitable therapeutic target to suppress DC activation in inflammation or autoimmunity.
Keywords: Dendritic cell activation, plasmacytoid dendritic cell, conventional dendritic cell, etomoxir, TOFA, 5-(Tetradecyloxy)-2-furoic Acid, fatty acid oxidation, fatty acid synthase, systemic lupus erythematosus, pro-inflammatory cytokines, immunometabolism
Introduction
Dendritic cells (DCs) comprise a small percentage of the total population of immune cells, but their powerful capacity to produce pro- and anti-inflammatory cytokines and present antigens to T cells grants them the ability to shape an immune response.[1–3] While this is beneficial in a healthy immune system, overactive DCs have been implicated in the pathogenesis of many autoimmune diseases.[4–7] Of these, plasmacytoid dendritic cells (pDCs), the primary producer of the family of cytokines type I IFNs, are considered important in systemic lupus erythematosus (SLE),[8, 9] a complex systemic autoimmune disease in which the majority of patients show increased expression of type I IFN-stimulated genes (ISGs).[10–12] This immune dysregulation is seen in up to 80% of SLE patients and is termed the Interferon Signature.[10, 13] While pDCs are major producers of IFNα, conventional DCs (cDCs) produce mostly IFNβ, together with many other pro-inflammatory cytokines such as IL-6, and DC subsets have been found abnormally activated in lupus autoimmunity.[14–16]
The Lupus Foundation of America estimates that at least five million people worldwide—including 1.5 million Americans—have a form of lupus.[17] However, the precise etiology of SLE initiation and flares are poorly understood, thus targeted therapies for this disease are presently insufficient. Current treatments in SLE include antimalarials and systemic corticosteroids, which induce general immunosuppression and manage symptoms but do not often halt disease progression.[18] Recent clinical trials inhibiting IFNα have also failed, and it is evident that a full dissection of the aberrant operations of key immune players in SLE is needed to understand the processes that govern pathogenesis, in order to design interventional treatments that can affect disease progression.[19] As we begin to link metabolism with immune activation, metabolic manipulation of key players in SLE is emerging as a promising new target.[20]
Recent studies have shown that changes in the way in which immune cells produce their energy is important for their functions[21, 22] and is a putative therapeutic target in autoimmunity.[23] In particular, fatty acid (FA) metabolism has been proposed to be essential for pDC activation and response to type I IFNs.[24] Specifically, type I IFNs have been shown to promote both fatty acid oxidation (FAO) and oxidative phosphorylation (OXPHOS). In Toll-like receptor 9 (TLR9) agonist CpGA-stimulated pDCs, the enhanced FAO due to autocrine type I IFN signaling was found to be critical for pDC activation. Inhibition of this metabolic reprogramming significantly impaired full pDC activation while having no effect on cell viability.[24] The role of FAO and fatty acid synthesis (FAS) in cDCs, which can be generated in culture from murine bone marrow precursors in presence of FLT3-L, was not investigated. On the contrary, bone marrow-derived cDCs generated in culture in presence of GM-CSF and considered a murine model of proinflammatory DCs (iDCs), have been shown to upregulate glycolysis and shutdown oxidative phosphorylation upon stimulation, and require mostly glycolysis to sustain activation and their Ag presentation.[25, 26] In this study, we investigated the effects of the pharmacological blockade of FAO and FAS to manipulate pDC and cDC activation. This may be a safer and more efficient approach in autoimmune diseases that are mediated by high expression of type I IFNs, as targeting pDC and cDCs output may spare the response to type I IFN in other cell types.
Among the current pharmacologic tools used to manipulate cell metabolism, we investigated etomoxir, an irreversible inhibitor of carnitine palmitoyltransferase-1 (CPT-1), a key enzyme in FAO, critical transporter of long chain fatty acids into mitochondria for oxidation.[27] Additionally, we investigated the effects of 5-tetradecyloxy-2-furoic acid (TOFA), an inhibitor of acetyl-CoA carboxylase-α (ACC), targeting the rate-limiting enzyme in long-chain fatty acid synthesis. Dendritic cell dysfunction in cancer has been associated with lipid accumulation, and administration of TOFA has been found to normalize DC functional activity.[28, 29] We explored how fatty acid metabolism may be essential for DC activation and production of proinflammatory cytokines. We show that inhibiting FA metabolism by etomoxir and TOFA suppresses both pDC and cDC activation, suggesting these drugs as novel therapeutics for affecting DCs.
Materials & Methods
Mice
C57BL/6 (B6) wildtype mice were bred and maintained in our colony. Breeding pairs were originally purchased from Jackson Laboratory. Mice were between 8 and 12 weeks of age when used for experiments. Mice were housed in the Laboratory Animal Resources Unit of Temple University School of Medicine, an AALAC accredited facility. All experimental procedures reported were approved by the Temple University Institutional Animal Care and Use Committee (IACUC).
In Vitro Dendritic Cell Studies
Bone marrow derived pDCs and cDCs were generated as per the laboratory’s published protocols.[16, 26, 30–33] Briefly, bone marrow precursors were flushed from femurs and tibias of mice and differentiated into pDCs and cDCs in the presence of complete RPMI medium (Thermo Fisher), containing 10% FBS (Gemini Bio Products), L-glutamine (VWR), penicillin/streptomycin (VWR), 2-mercaptoethanol (Gibco, Thermo Fisher), and enriched with 15% FLT3-L conditioned medium from a FLT3-L-producing cell line, in 24 well plates for 7 days.[16] DCs were then sorted using the EasySep PE positive selection kit (StemCell Technologies) with a PE-labelled anti-B220 antibody (StemCell Technologies) to separate out the pDCs from the cDCs. Purity of cells averaged at least 90%. Alternatively, pDCs and cDCs were purified by FACS, reaching a purity >99%. Sorted cells were plated in complete RPMI medium with FLT3-L over-night, then stimulated the next morning. TLR9 ligands CpGA 5ug/ml (ODN 2336 synthesized by IDT Biotechnologies) was used to stimulate both pDCs and cDCs. R-(+)-Etomoxir and TOFA were both purchased from Cayman Chemical and used at varying concentration, adding them 1 hour before the CpG-A stimulation. 24 hours post-stimulation, supernatants were collected for measurement of cytokine production and cells were harvested by gentle pipetting before to be stained with specific antibodies for analysis by flow cytometry.
Cytokine ELISA
IL-6 was measured in the supernatants of pDC and cDC cultures using the BD OptEIA™ Mouse IL-6 ELISA kit, and CXCL10 using the R&D Systems Mouse CXCL10/IP-10/CRG-2 DuoSet ELISA kit. Optical densities were measured at 650 nm and 405 nm according to the manufacturer’s protocol and results analyzed with SoftMax Pro software (Molecular Devices Corporation, Sunnyvale, CA).
Flow Cytometry
Following harvesting, pDCs and cDCs were incubated with rat anti-mouse CD16/CD32 mAb (Biolegend) for 15 min on ice to block FcγRs. Cells were then stained for 30 min in the dark on ice using specific antibodies for surface identification markers CD11c (eBioscience), CD11b (BD Bioscience), and B220 (StemCell Technologies), and costimulatory marker CD86 and CD40 (BD Bioscience). Cells were analyzed for cell viability using Fixable Viability Dye eFluor 780 (Thermo Fisher). Cells were fixed in 2% paraformaldehyde (Thomas Scientific) in PBS (Fisher) with 1% BSA (Gemini Bio Products). All cells were analyzed on a FACSCanto flow cytometer (BD Bioscience) with FlowJo software (Tree Star, Ashland, OR, USA).
Statistical Analysis
Prism 8 (GraphPad software, San Diego, CA, USA) was used for data analysis. Means and standard error of means (Mean ± SEM) were calculated by averaging results from independent experiments. Statistical significance was determined using one-way ANOVA and multiple comparisons post-hoc correction test. P-values marked in the figures as * P<0.05, ** P<0.01, *** P<0.001 and **** P<0.0001 were considered significant.
Results
Inhibition of fatty acid oxidation by etomoxir suppresses up-regulation of costimulatory molecules in pDCs and cDCs
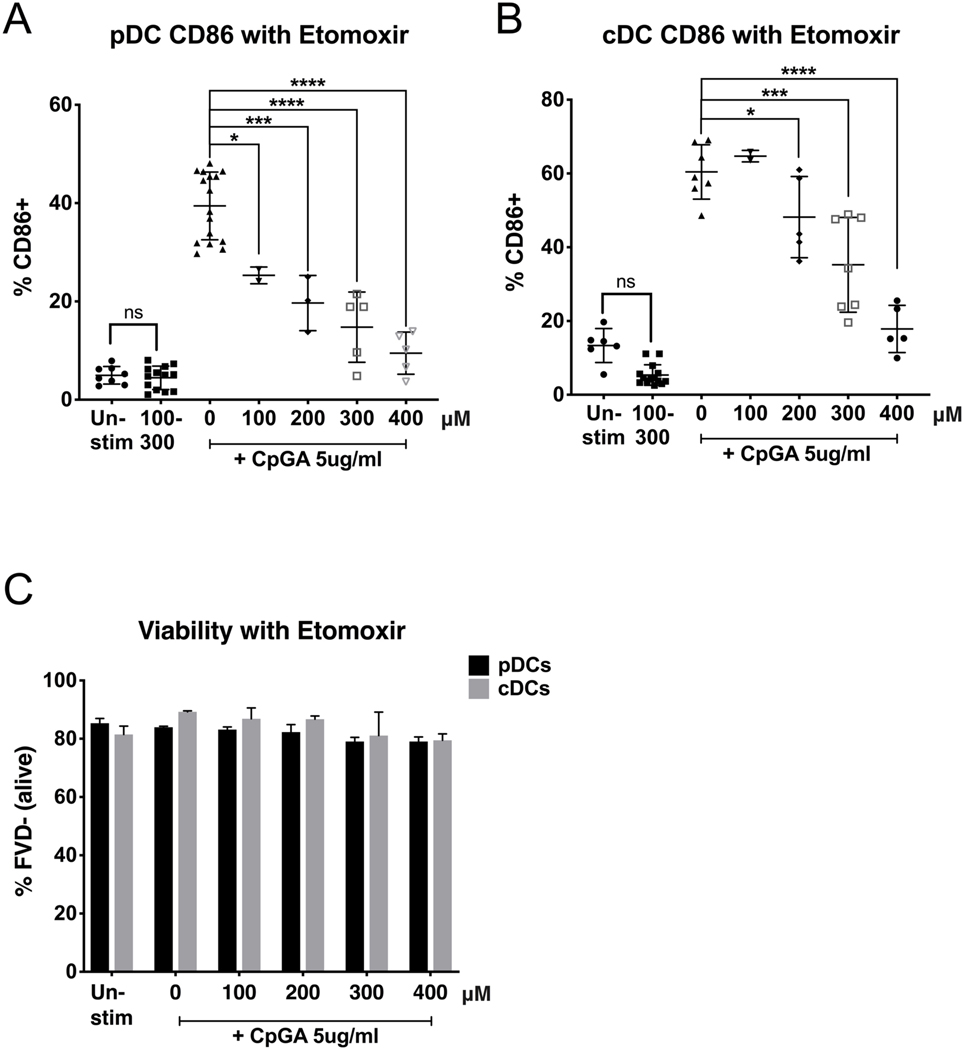
As sentinels of the immune system, DCs are relatively quiescent at the steady state, but are able to very rapidly respond to perturbations. To determine the role of fatty acid metabolism in dendritic cell activation, we grew bone marrow precursors in presence of FLT3-L, a well establish protocol that elicits a mixed population of pDCs and cDCs. After sorting with magnetic beads, we obtained single subsets of pDCs and cDCs, with a purity of at least 90% (Figure 1). Similar experiments were repeated using pDCs and cDCs that were purified by Flow Sorting, reaching a purity >99%. After an over-night rest, we treated pDCs and cDCs with a dose titration of etomoxir, an inhibitor of the key fatty acid oxidation (FAO) enzyme carnitine palmitoyltransferase I (CPT1). We chose this dose range following previous literature in DCs and T cells.[24, 34, 35] After 1 hour, we stimulated both DC subsets with TLR9 ligand CpG-A 2336, an oligonucleotide that is able to stimulate both subsets of murine DCs (Figure 2). We found that etomoxir suppressed both pDC and cDC activation as measured by the expression of surface co-stimulatory molecule CD86 (Figure 2a and b). Etomoxir suppressed TLR-induced CD86 up-regulation of both DC subsets in a dose-dependent manner, and pDCs were sensitive to inhibition at lower concentrations (100μM) than cDCs (200μM). In both DC subsets, etomoxir did not affect cell viability, neither in resting state or upon stimulation (Figure 2c), suggesting that suppressing FAO inhibits the metabolic switching needed for activation, without inducing cell death in resting nor stimulated DCs. Etomoxir treatment did not affect the mean fluorescence intensity (MFI) of CD11c, CD11b, and B220 differentiation markers and subset percentages, suggesting that it does not affect DC differentiation (not shown).
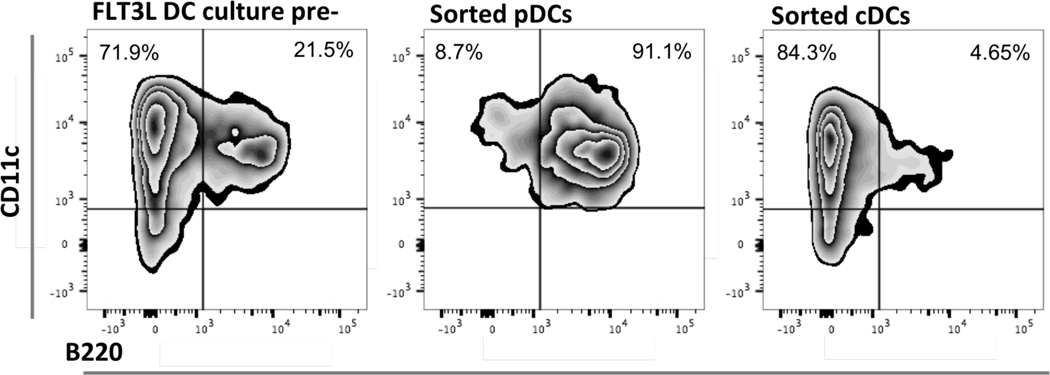
Figure 1. Sorting FLT3-L DC culture by positive selection of pDCs using surface expression of B220.
pDCs and cDCs were generated in vitro from bone marrow precursor cells harvested from the femur and tibia of WT mice in FLT3-L enriched-medium. After 7 days in culture, cells were gently harvested, and pDCs were positively selected for surface expression of B220, a CD45 isoform expressed by murine pDCs but not cDCs. The positively selected cohort of cells for all experiments displayed a purity of at least 90%. Flow diagrams are representative of purity checks for all the experiments in which pDCs were sorted by magnetic beads, showing the expression of CD11c and B220 before sorting (left) and in the positively selected pDC subset (center) and cDC subset (right).
Figure 2. Inhibition of fatty acid oxidation by etomoxir suppresses DC upregulation of CD86.
Purified pDCs and cDCs were treated for 1h with CPT1 inhibitor etomoxir (concentrations reported in μM), then stimulated with TLR9 agonist CpGA 5μg/ml. Activation was measured after 24h by CD86 expression via flow cytometry. pDCs were gated for B220+CD11c+CD11b-. cDCs were gated for B220-CD11c+CD11b+. Viability of cells (C) is expressed as % of cells negative for the Fixable Viability Dye and was not affected at effective concentrations of etomoxir. Results are shown as the mean ± SEM, from 5 independent experiments. Significance was calculated by one-way ANOVA and multiple comparisons post-hoc correction test, and P-values marked in the figures as * P<0.05, ** P<0.01, *** P<0.001 and **** P<0.0001 were considered significant.
pDCs have a lower threshold of sensitivity to etomoxir than cDCs
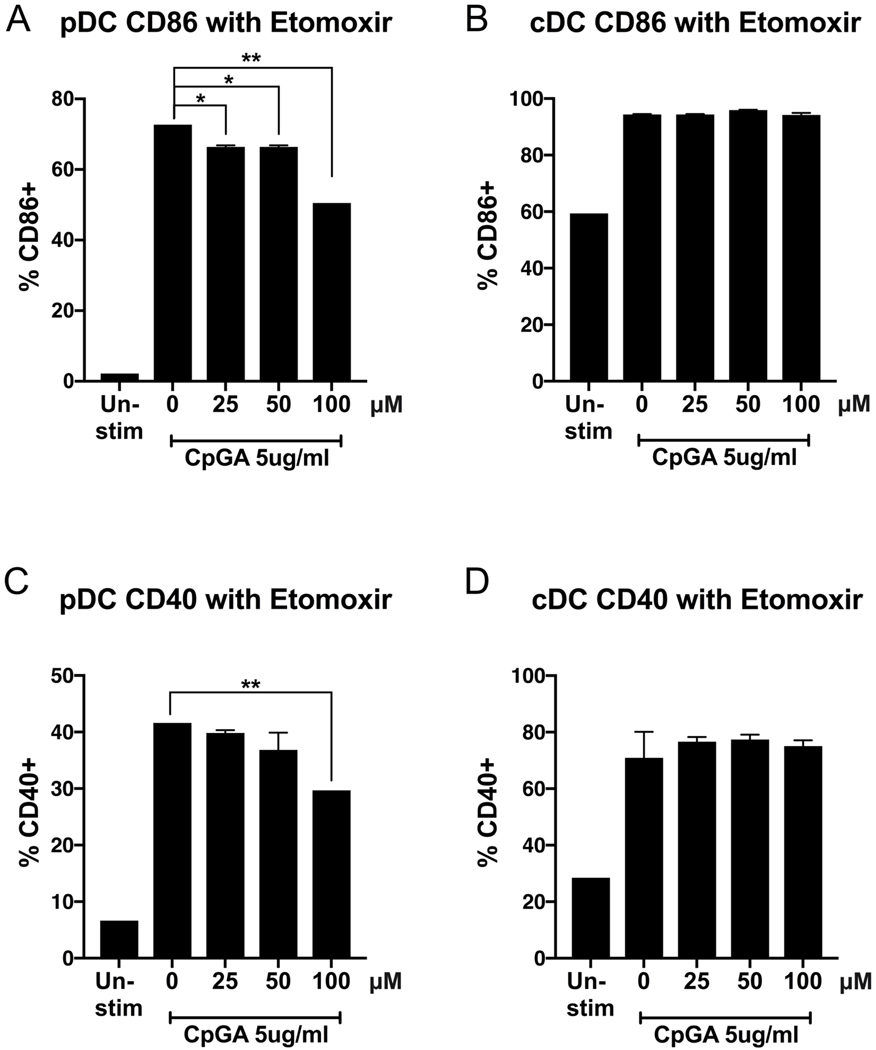
The dose titration shown in Figure 2 did not reveal the threshold of sensitivity of pDCs to etomoxir. Therefore, we further tested lower concentrations of the inhibitor. Since magnetic bead-purified pDC and cDC populations reached a high purity of at least 90%, we confirmed our results and tested lower doses of etomoxir on an ultrapure cell population. Sorting with BD FACSAria IIμ, we obtained 99.9% pure populations of FLT3-L-derived bone marrow pDCs and cDCs that we treated with etomoxir, ranging from 25μM up to 100μM, the bead-sorted lowest dose used in Figure 2. After 1 hour of treatment, both DC subsets were stimulated with CpGA. After 24 hours, cells were harvested for flow cytometry analysis of surface costimulatory molecule CD86 and CD40. Etomoxir suppressed the TLR-induced CD86 upregulation by pDCs at concentrations as low as 25μM. Etomoxir-induced suppression of pDC upregulation of CD40 exhibited a similar trend as seen with CD86, though the suppression reached significance only at 100μM (Figures 3a and 3c), suggesting distinct requirements of activation for the expression of these costimulatory molecules. cDCs were not sensitive to inhibition at these lower concentrations of etomoxir (Figures 3b and 3d), confirming the results shown in Figure 2. Viability of these ultrapure populations was not affected by etomoxir (not shown). This data shows that suppressing FAO by etomoxir affects pDCs at lower concentrations than cDCs, suggesting that TLR-induced pDC activation may be more susceptible to fatty acid metabolic modulation than cDCs.
Figure 3. Low concentrations of Etomoxir suppress the upregulation of costimulatory molecule CD86 and CD40 in pDC but not cDC subsets.
Flow cytometry sorted ultrapure pDC (A, C) and cDC (B, D) populations were treated for 1h with etomoxir (25–100 μM), then stimulated with TLR9 agonist CpGA 5μg/ml. Activation was measured after 24h by CD86 and CD40 expression via flow cytometry. pDCs were gated for B220+CD11c+CD11b-. cDCs were gated for B220-CD11c+CD11b+. Results are shown as the mean ± SEM of biological triplicates. Significance was calculated by one-way ANOVA and multiple comparisons post-hoc correction test, and P-values marked in the figures as * P<0.05, ** P<0.01, *** P<0.001 and **** P<0.0001 were considered significant.
Inhibition of fatty acid synthesis by TOFA suppresses DC activation
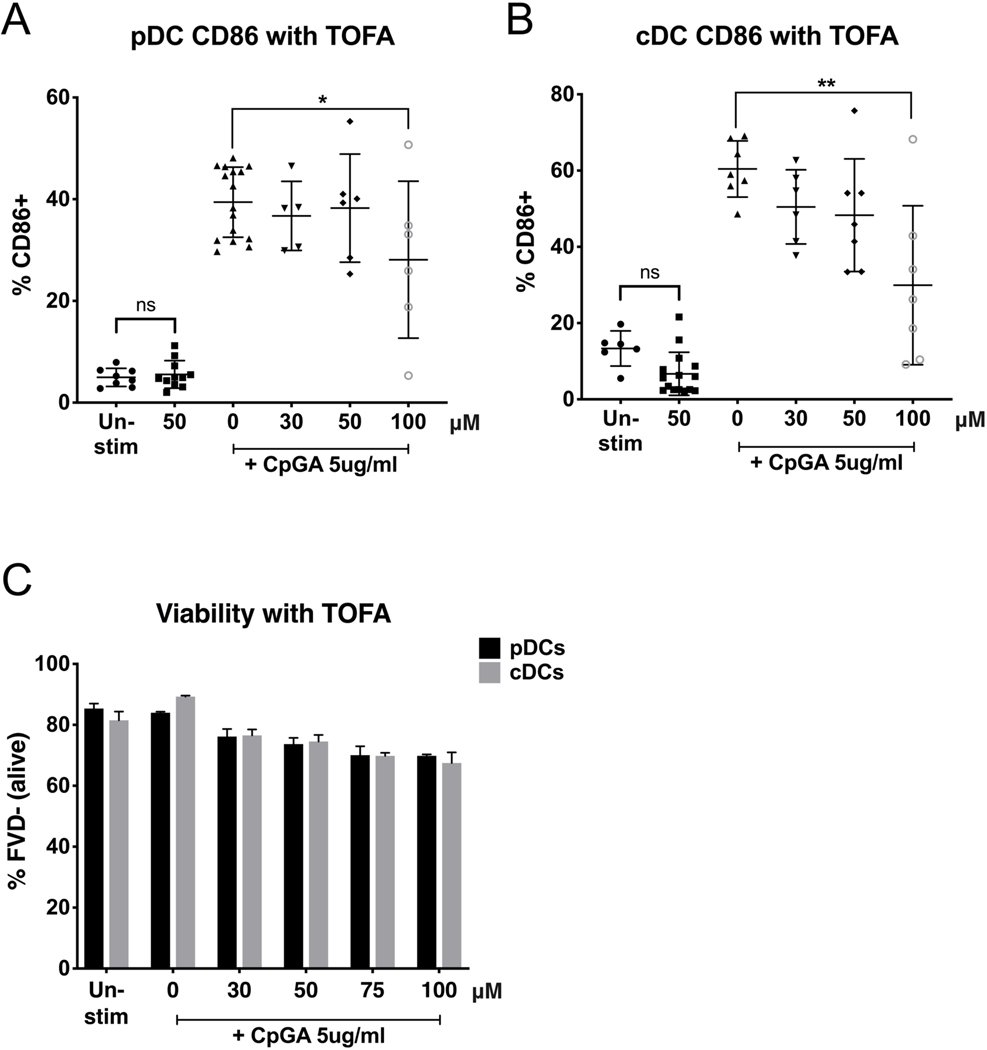
Next, we investigated whether inhibiting fatty acid synthesis (FAS), which may also contribute to FAO in DCs, would also affect activation. We treated magnetic bead-purified pDCs and cDCs with a dose titration of 5-tetradecyloxy-2-furoic acid, or TOFA, an inhibitor of acetyl-CoA carboxylase-α (ACC), using doses previous tested in immune cells.[24, 29] We found that TOFA did not affect cell viability (Figure 4c), neither in resting state nor upon stimulation, but inhibited the upregulation of surface co-stimulatory molecule CD86 induced by CpG-A in both DC subsets at the same concentration of 100μM (Figures 4a and 4b). As for etomoxir, these findings suggest that TOFA is effective in inhibiting the metabolic switch needed for pDCs and cDCs to activate without shutting down the metabolic pathways that are keeping them alive.
Figure 4. Inhibition of fatty acid synthesis by TOFA suppresses pDC and cDC upregulation of CD86.
Purified pDCs (A) and cDCs (B) were treated for 1h with TOFA (doses expressed in μM), an inhibitor of acetyl-CoA carboxylase-α, then stimulated with TLR9 agonist CpGA 5μg/ml. Activation was measured after 24h by CD86 expression via flow cytometry. pDCs were gated for B220+CD11c+CD11b-. cDCs were gated for B220-CD11c+CD11b+. Viability of cells (C) is expressed as % of cells negative for the Fixable Viability Dye. Results are shown as the mean ± SEM, from 5 independent experiments. Significance was calculated by one-way ANOVA and multiple comparisons post-hoc correction test, and P-values marked in the figures as * P<0.05, ** P<0.01, *** P<0.001 and **** P<0.0001 were considered significant.
Inhibition of fatty acid metabolism suppresses DC production of proinflammatory cytokine IL-6
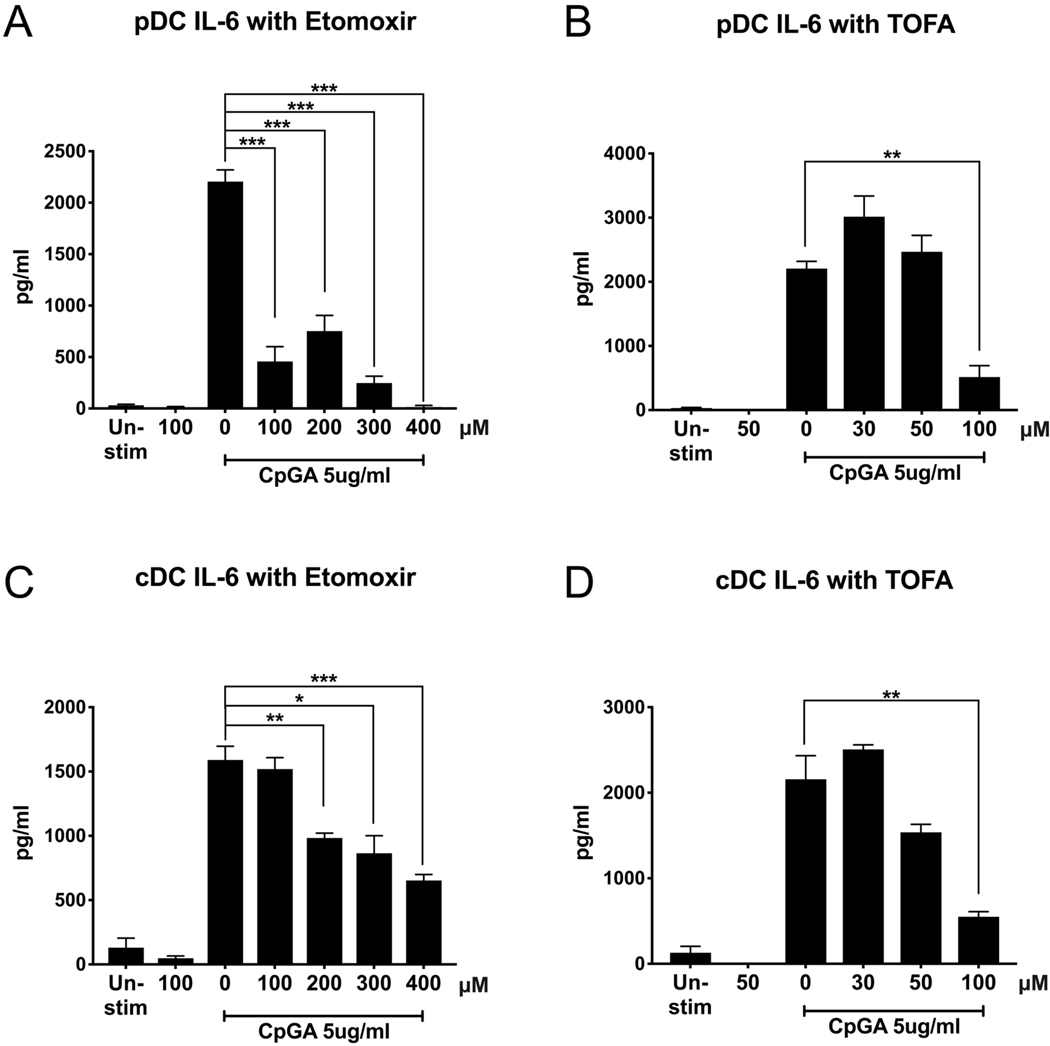
To further study the role of FA metabolism on DC activation, we next investigated whether inhibiting FA metabolism affects the production of proinflammatory cytokines by DCs. Notably, IL-6 production is found to be higher in lupus patients and has been shown to reflect disease activity, including autoantibody titers. [36–39] Testing the supernatants of pDCs and cDCs treated with etomoxir to inhibit FAO and TOFA to inhibit FAS, as described above, we found that the inhibition of FA metabolism is sufficient to suppress the production of proinflammatory cytokine interleukin (IL)-6 by pDCs (Figure 5a and 5b), and by cDCs (Figure 5c and 5d). This finding suggests that etomoxir and TOFA are able to fully suppress DC proinflammatory response, while still allowing both pDCs and cDCs to maintain baseline metabolism, preserving their viability.
Figure 5. Metabolic modulation of fatty acid metabolism suppresses proinflammatory IL-6 production.
Purified pDCs and cDCs were treated for 1h with FAO inhibitor etomoxir (μM) or FAS inhibitor TOFA(μM), then stimulated with TLR9 agonist CpGA 5μg/ml. Supernatant was collected after 24h and IL-6 production was measured by ELISA. Results are shown as the mean ± SEM, from biological triplicate of 2 independent experiments. Significance was calculated by one-way ANOVA and multiple comparisons post-hoc correction test, and P-values marked in the figures as * P<0.05, ** P<0.01, *** P<0.001 and **** P<0.0001 were considered significant.
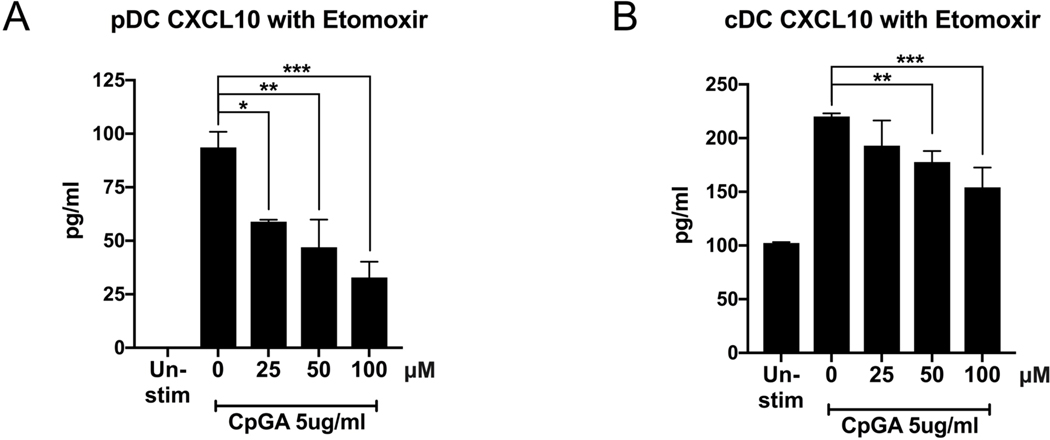
Production of type I IFN-dependent chemokine CXCL10 is suppressed by etomoxir
Type I IFNs have been implicated in regulating the metabolic changes necessary for pDC activation.[24] Therefore, we next investigated whether modulation of fatty acid metabolism affected the production of the chemokine CXCL10, an Interferon Stimulated gene, which we chose as a measurement of DC response to autocrine type I IFNs because we have previously shown that this ckemokine is induced in DCs by TLR stimulation in a type I IFN dependent manner.[31, 40] Supernatants from pDCs and cDCs, treated with etomoxir as in Figure 3, showed a statistically significant suppression of the production of ISG CXCL10 in response to CpGA stimulation (Figure 6a), consistent with Figure 3 data showing that low concentrations of etomoxir are able to suppress pDC upregulation of CD86. Interestingly, although etomoxir did not affect cDC upregulation of surface costimulatory molecules until concentrations of at least 200μM, etomoxir as low as 50μM significantly suppressed cDC production of CXCL10 (Figure 6b). This finding suggests that modulation of FAO by etomoxir may suppress the type I IFN response at concentrations lower than those that affect the expression of surface costimulatory molecules.
Figure 6. Metabolic modulation of fatty acid oxidation by etomoxir suppresses type I IFN-induced CXCL10.
Flow cytometry sorted pDC (A) and cDC (B) populations were treated for 1h with etomoxir (μM), then stimulated with TLR9 agonist CpGA 5μg/ml. Supernatant was collected after 24h and analyzed for type I IFN-induced CXCL10 production by ELISA. Results are shown as the mean ± SEM of biological triplicates. Significance was calculated by one-way ANOVA and multiple comparisons post-hoc correction test, and P-values marked in the figures as * P<0.05, ** P<0.01, *** P<0.001 and **** P<0.0001 were considered significant.
Discussion
The immune system is comprised of cells that are relatively quiescent in the steady state, but rapidly respond to perturbations.[41] DCs act as the bridge between the innate and adaptive immune systems and, through presentation to T cells and production of cytokines, they direct the immune response.[42] Changes in energy pathways are necessary to mount an immune response.[22, 41, 43] Indeed, both innate cells and T lymphocytes rely on the process of mitochondrial oxidative phosphorylation to obtain energy when they are quiescent, while they increase their metabolic capacity and shift to glycolysis and fatty acid metabolism upon activation.[41] DCs are a diverse population comprising many subsets, with different roles and functions, and this diversity is associated with different metabolic requirements.
Our findings confirm that FA metabolism is important for TLR-induced activation of pDCs, as previously reported[22, 24]. Moreover, we show for the first time that FLT3-L-derived cDCs also activate with a similar requirement for FAO and FAS. These results suggest that FLT3/-L-cDCs have different energy requirements than GM-CSF-iDCs, known to shut down oxidative phosphorylation and rely on glycolysis after activation, and therefore it may be possible to therapeutically target one subset and not the other. By using FAO inhibitor etomoxir and FAS inhibitor TOFA, we suppressed the TLR-induced upregulation of surface costimulatory molecule CD86 and CD40, which are important for DCs to interact with T cells to mount an adaptive immune response. Additionally, the overexpression of CD86 in DCs from lupus patients suggests that manipulating CD86 could be a clinically relevant response.[44] Furthermore, etomoxir and TOFA suppressed the TLR-induced production of IL-6, a proinflammatory cytokine found to be produced in excess by stimulated DCs from lupus patients.[45, 46] and proposed to be an accurate biomarker of SLE disease activity. [39] The feasibility of inhibiting the upregulation of these markers of pDC and cDC activation with FAO and FAS inhibitors suggests a therapeutic potential for these drugs in lupus.
Moreover, we have found that etomoxir can inhibit the production of chemokine CXCL10, which we have previously shown to be an ISG produced by DCs upon TLR stimulation in a manner dependent on autocrine type I IFNs[31] suggesting that etomoxir can suppress the response of both pDCs and cDCs to type I IFNs. Type I IFNs have been shown to regulate the immunometabolism of pDCs upon TLR9 stimulation and increase FAO and FAS.[24] Our results extend these findings to FLT3-L cDCs, indicating that it is a general pathway for cDCs and pDCs to require active FA metabolism to respond to autocrine type I IFNs and complete DC activation.
We have recently reported that iDCs from lupus-prone mice have a higher energy metabolism than WT iDCs,[40] and it will be important to determine whether also cDCs and pDCs from lupus-prone mice have higher metabolism, and in particular higher FAO and FAS, and the interdependence between FA metabolism and the lupus IFN Signature. Serum levels of CXCL10 are elevated in SLE patients over controls[47], and more recently, CXCL10 has also been identified to have predictive value in SLE.[48]. Our results that FAO inhibition suppresses CXCL10 up-regulation suggest that FAO and/or FAS inhibition may normalize DC abnormalities in lupus.
We used concentrations of etomoxir and TOFA that were suggested by the literature and shown feasible in murine in vivo studies [34, 49–51]. As for many other inhibitors, these compounds can have off-target effects at high doses. In particular, it has been recently proposed in T cells and macrophages that etomoxir at the concentration of 3μM has no effects on intact cells, but at 200μM has definite off-target effects, namely suppression of OXPHOS through inhibition of complex I in the electron transport chain and through an adenine nucleotide translocase blockade.[52–54] The data leave a large window of efficacy that is specific for FAO inhibition. We found that etomoxir inhibited pDC activation and ISG production at 25μM, a concentration in range for specific inhibition of CPT-1. In cDCs, etomoxir inhibited the ISG production at 50μM and costimulatory molecules at 100μM, the latter a border line concentration that suggests that the effects of etomoxir on cDCs may be in part CPT-1 specific and in part due to off-target effects on OXPHOS. Since this window of sensitivity was reported for macrophages[54] and not in cDCs nor pDCs, it will be important to investigate the activation levels of FAO in these specific cell subsets in order to clarify the molecular targets of etomoxir in DCs. Ultimately, our results indicate that pDCs are more susceptible to etomoxir than cDCs, and in cDCs the response to type I IFN is more susceptible than the upregulation of costimulatory molecules. These results suggest that etomoxir in vivo may be more specific to inhibit responses to type I IFNs, such as the IFN Signature in autoimmunity.
Pharmacologic inhibition of FAO with etomoxir or FAS with TOFA has been reported inhibiting proliferation and sensitizing to cell death human tumor cells, like leukemia cells[55] and glioblastoma cells.[27] Therefore, we analyzed a possible effect of these drugs on DC survival. The results that both drugs inhibit metabolic reprogramming to a degree that suppresses DC activation without affecting cell viability, indicate that FA metabolism is not required for the survival of resting pDCs and cDCs during homeostatic conditions. Moreover, they suggest that FAO and FAS may be an exploitable target for novel therapies to suppress DC activation in inflammation or autoimmunity because inhibition of FA metabolism may suppress DC activation, and therefore immunity, without killing the resting nor the tolerogenic DCs.
In conclusion, we show that modulation of FAO by etomoxir may be more effective than modulation of FAS by TOFA for suppressing the activation of both pDCs and cDCs. Our data also show that low doses of etomoxir can significantly inhibit the production of type I IFN-stimulated genes by both pDCs and cDCs, emphasizing how shifts in metabolism are important for immune cell activation, and highlighting FAO as a specific pathway important for the activation of two main subsets of DCs. A surge in immunometabolism research has led to increased interest in metabolic modulators, and their potential as novel therapeutics. Our studies show how a range of etomoxir and TOFA doses affects pDC and cDC outcomes of activation, related to different immune responses, suggesting the possibility to modulate tolerance vs specific immune responses, rather the blindly inhibit the immune system, like present immunosuppressive therapies do. In summary, our findings suggest that DC metabolism may be an exploitable target in SLE.
Acknowledgement
We would like to thank the Temple Infections and Autoimmunity Interest Group for stimulating discussions.
Funding:
This work was supported by the U.S, National Institutes of Health, NIAD, R21 AI119947 (SG), and the Lupus Foundation of America, under the Goldie Simon Preceptorship Award (CCQ).
Footnotes
Declaration of interest/disclosure
No financial benefit has arisen from the findings in this research. No conflicts of interest are reported by any of the authors.
References
- [1].Merad M, Sathe P, Helft J, Miller J, and Mortha A, The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting, Annu Rev Immunol 31 (2013), pp. 563–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Guilliams M, Ginhoux F, Jakubzick C, Naik SH, Onai N, Schraml BU, Segura E, Tussiwand R, and Yona S, Dendritic cells, monocytes and macrophages: a unified nomenclature based on ontogeny, Nat Rev Immunol 14 (2014), pp. 571–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Gallo PM, and Gallucci S, The dendritic cell response to classic, emerging, and homeostatic danger signals. Implications for autoimmunity, Front Immunol 4 (2013), p. 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ganguly D, Haak S, Sisirak V, and Reizis B, The role of dendritic cells in autoimmunity, Nat Rev Immunol 13 (2013), pp. 566–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Quintana FJ, Dendritic cells in autoimmunity, infections, and cancer, Semin Immunopathol 39 (2017), pp. 97–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Mbongue J, Nicholas D, Firek A, and Langridge W, The role of dendritic cells in tissue-specific autoimmunity, J Immunol Res 2014 (2014), p. 857143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Drakesmith H, Chain B, and Beverley P, How can dendritic cells cause autoimmune disease?, Immunol Today 21 (2000), pp. 214–7. [DOI] [PubMed] [Google Scholar]
- [8].Amodio G, and Gregori S, Dendritic cells a double-edge sword in autoimmune responses, Front Immunol 3 (2012), p. 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hardin JA, Dendritic cells: potential triggers of autoimmunity and targets for therapy, Ann Rheum Dis 64 Suppl 4 (2005), pp. iv86–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Crow MK, Type I interferon in the pathogenesis of lupus, J Immunol 192 (2014), pp. 5459–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Rönnblom L, Alm GV, and Eloranta ML, Type I interferon and lupus, Curr Opin Rheumatol 21 (2009), pp. 471–7. [DOI] [PubMed] [Google Scholar]
- [12].Bennett L, Palucka AK, Arce E, Cantrell V, Borvak J, Banchereau J, and Pascual V, Interferon and granulopoiesis signatures in systemic lupus erythematosus blood, J Exp Med 197 (2003), pp. 711–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Obermoser G, and Pascual V, The interferon-alpha signature of systemic lupus erythematosus, Lupus 19 (2010), pp. 1012–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Guiducci C, Gong M, Xu Z, Gill M, Chaussabel D, Meeker T, Chan JH, Wright T, Punaro M, Bolland S, Soumelis V, Banchereau J, Coffman RL, Pascual V, and Barrat FJ, TLR recognition of self nucleic acids hampers glucocorticoid activity in lupus, Nature 465 (2010), pp. 937–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Colonna L, Dinnall JA, Shivers DK, Frisoni L, Caricchio R, and Gallucci S, Abnormal costimulatory phenotype and function of dendritic cells before and after the onset of severe murine lupus, Arthritis Res Ther 8 (2006), p. R49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Sriram U, Varghese L, Bennett HL, Jog NR, Shivers DK, Ning Y, Behrens EM, Caricchio R, and Gallucci S, Myeloid dendritic cells from B6.NZM Sle1/Sle2/Sle3 lupus-prone mice express an IFN signature that precedes disease onset, J Immunol 189 (2012), pp. 80–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Statistics on Lupus, Lupus Foundation of America, 2016. [Google Scholar]
- [18].Kaul A, Gordon C, Crow MK, Touma Z, Urowitz MB, van Vollenhoven R, Ruiz-Irastorza G, and Hughes G, Systemic lupus erythematosus, Nat Rev Dis Primers 2 (2016), p. 16039. [DOI] [PubMed] [Google Scholar]
- [19].Dolgin E, Lupus in crisis: as failures pile up, clinicians call for new tools, Nat Biotechnol 37 (2019), pp. 7–8. [DOI] [PubMed] [Google Scholar]
- [20].Perl A, Review: Metabolic Control of Immune System Activation in Rheumatic Diseases, Arthritis Rheumatol 69 (2017), pp. 2259–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Geltink RIK, Kyle RL, and Pearce EL, Unraveling the Complex Interplay Between T Cell Metabolism and Function, Annu Rev Immunol 36 (2018), pp. 461–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].O’Neill LA, and Pearce EJ, Immunometabolism governs dendritic cell and macrophage function, J Exp Med 213 (2016), pp. 15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Yin Y, Choi SC, Xu Z, Perry DJ, Seay H, Croker BP, Sobel ES, Brusko TM, and Morel L, Normalization of CD4+ T cell metabolism reverses lupus, Sci Transl Med 7 (2015), p. 274ra18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wu D, Sanin DE, Everts B, Chen Q, Qiu J, Buck MD, Patterson A, Smith AM, Chang CH, Liu Z, Artyomov MN, Pearce EL, Cella M, and Pearce EJ, Type 1 Interferons Induce Changes in Core Metabolism that Are Critical for Immune Function, Immunity 44 (2016), pp. 1325–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Everts B, Amiel E, van der Windt GJ, Freitas TC, Chott R, Yarasheski KE, Pearce EL, and Pearce EJ, Commitment to glycolysis sustains survival of NO-producing inflammatory dendritic cells, Blood 120 (2012), pp. 1422–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Chakhtoura M, Chain RW, Sato PY, Qiu CC, Lee MH, Meissler JJ, Eisenstein TK, Koch WJ, Caricchio R, and Gallucci S, Ethyl Pyruvate Modulates Murine Dendritic Cell Activation and Survival Through Their Immunometabolism, Front Immunol 10 (2019), p. 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Pike LS, Smift AL, Croteau NJ, Ferrick DA, and Wu M, Inhibition of fatty acid oxidation by etomoxir impairs NADPH production and increases reactive oxygen species resulting in ATP depletion and cell death in human glioblastoma cells, Biochim Biophys Acta 1807 (2011), pp. 726–34. [DOI] [PubMed] [Google Scholar]
- [28].Guseva NV, Rokhlin OW, Glover RA, and Cohen MB, TOFA (5-tetradecyl-oxy-2-furoic acid) reduces fatty acid synthesis, inhibits expression of AR, neuropilin-1 and Mcl-1 and kills prostate cancer cells independent of p53 status, Cancer Biol Ther 12 (2011), pp. 80–5. [DOI] [PubMed] [Google Scholar]
- [29].Herber DL, Cao W, Nefedova Y, Novitskiy SV, Nagaraj S, Tyurin VA, Corzo A, Cho HI, Celis E, Lennox B, Knight SC, Padhya T, McCaffrey TV, McCaffrey JC, Antonia S, Fishman M, Ferris RL, Kagan VE, and Gabrilovich DI, Lipid accumulation and dendritic cell dysfunction in cancer, Nat Med 16 (2010), pp. 880–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Gallo PM, Rapsinski GJ, Wilson RP, Oppong GO, Sriram U, Goulian M, Buttaro B, Caricchio R, Gallucci S, and Tükel Ç, Amyloid-DNA Composites of Bacterial Biofilms Stimulate Autoimmunity, Immunity 42 (2015), pp. 1171–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Xu J, Lee MH, Chakhtoura M, Green BL, Kotredes KP, Chain RW, Sriram U, Gamero AM, and Gallucci S, STAT2 Is Required for TLR-Induced Murine Dendritic Cell Activation and Cross-Presentation, J Immunol 197 (2016), pp. 326–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sriram U, Biswas C, Behrens EM, Dinnall JA, Shivers DK, Monestier M, Argon Y, and Gallucci S, IL-4 suppresses dendritic cell response to type I interferons, J Immunol 179 (2007), pp. 6446–55. [DOI] [PubMed] [Google Scholar]
- [33].Sriram U, Xu J, Chain RW, Varghese L, Chakhtoura M, Bennett HL, Zoltick PW, and Gallucci S, IL-4 suppresses the responses to TLR7 and TLR9 stimulation and increases the permissiveness to retroviral infection of murine conventional dendritic cells, PLoS One 9 (2014), p. e87668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Shriver LP, and Manchester M, Inhibition of fatty acid metabolism ameliorates disease activity in an animal model of multiple sclerosis, Sci Rep 1 (2011), p. 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hossain F, Al-Khami AA, Wyczechowska D, Hernandez C, Zheng L, Reiss K, Valle LD, Trillo-Tinoco J, Maj T, Zou W, Rodriguez PC, and Ochoa AC, Inhibition of Fatty Acid Oxidation Modulates Immunosuppressive Functions of Myeloid-Derived Suppressor Cells and Enhances Cancer Therapies, Cancer Immunol Res 3 (2015), pp. 1236–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zhu J, Liu X, Xie C, Yan M, Yu Y, Sobel ES, Wakeland EK, and Mohan C, T cell hyperactivity in lupus as a consequence of hyperstimulatory antigen-presenting cells, J Clin Invest 115 (2005), pp. 1869–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ding D, Mehta H, McCune WJ, and Kaplan MJ, Aberrant phenotype and function of myeloid dendritic cells in systemic lupus erythematosus, J Immunol 177 (2006), pp. 5878–89. [DOI] [PubMed] [Google Scholar]
- [38].Fransen JH, van der Vlag J, Ruben J, Adema GJ, Berden JH, and Hilbrands LB, The role of dendritic cells in the pathogenesis of systemic lupus erythematosus, Arthritis Res Ther 12 (2010), p. 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Abdel Galil SM, Ezzeldin N, and El-Boshy ME, The role of serum IL-17 and IL-6 as biomarkers of disease activity and predictors of remission in patients with lupus nephritis, Cytokine 76 (2015), pp. 280–287. [DOI] [PubMed] [Google Scholar]
- [40].Lee MH, Chakhtoura M, Sriram U, Caricchio R, and Gallucci S, Conventional DCs from Male and Female Lupus-Prone B6.NZM Sle1/Sle2/Sle3 Mice Express an IFN Signature and Have a Higher Immunometabolism That Are Enhanced by Estrogen, J Immunol Res 2018 (2018), p. 1601079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Pearce EL, and Pearce EJ, Metabolic pathways in immune cell activation and quiescence, Immunity 38 (2013), pp. 633–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Steinman RM, Linking innate to adaptive immunity through dendritic cells, Novartis Found Symp 279 (2006), pp. 101–9; discussion 109–13, 216–9. [PubMed] [Google Scholar]
- [43].Ganeshan K, and Chawla A, Metabolic regulation of immune responses, Annu Rev Immunol 32 (2014), pp. 609–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Decker P, Kötter I, Klein R, Berner B, and Rammensee HG, Monocyte-derived dendritic cells over-express CD86 in patients with systemic lupus erythematosus, Rheumatology (Oxford) 45 (2006), pp. 1087–95. [DOI] [PubMed] [Google Scholar]
- [45].Brahmakshatriya V, Kuang Y, Devarajan P, Xia J, Zhang W, Vong AM, and Swain SL, IL-6 Production by TLR-Activated APC Broadly Enhances Aged Cognate CD4 Helper and B Cell Antibody Responses In Vivo, J Immunol 198 (2017), pp. 2819–2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Ohl K, and Tenbrock K, Inflammatory cytokines in systemic lupus erythematosus, J Biomed Biotechnol 2011 (2011), p. 432595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Bauer JW, Baechler EC, Petri M, Batliwalla FM, Crawford D, Ortmann WA, Espe KJ, Li W, Patel DD, Gregersen PK, and Behrens TW, Elevated serum levels of interferon-regulated chemokines are biomarkers for active human systemic lupus erythematosus, PLoS Med 3 (2006), p. e491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Andrade F, Akhter E, Fang H, and Petri M, ABSTRACT NUMBER: 619 Elevated Plasma Levels of CXCL2 and CXCL10 Have Distinct Predictive Value in Systemic Lupus Erythematosus, American College of Rheumatology (2012). [Google Scholar]
- [49].Timmers S, Nabben M, Bosma M, van Bree B, Lenaers E, van Beurden D, Schaart G, Westerterp-Plantenga MS, Langhans W, Hesselink MK, Schrauwen-Hinderling VB, and Schrauwen P, Augmenting muscle diacylglycerol and triacylglycerol content by blocking fatty acid oxidation does not impede insulin sensitivity, Proc Natl Acad Sci U S A 109 (2012), pp. 11711–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Briant LJB, Dodd MS, Chibalina MV, Rorsman NJG, Johnson PRV, Carmeliet P, Rorsman P, and Knudsen JG, CPT1a-Dependent Long-Chain Fatty Acid Oxidation Contributes to Maintaining Glucagon Secretion from Pancreatic Islets, Cell Rep 23 (2018), pp. 3300–3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Ibitokou SA, Dillon BE, Sinha M, Szczesny B, Delgadillo A, Reda Abdelrahman D, Szabo C, Abu-Elheiga L, Porter C, Tuvdendorj D, and Stephens R, Early Inhibition of Fatty Acid Synthesis Reduces Generation of Memory Precursor Effector T Cells in Chronic Infection, J Immunol 200 (2018), pp. 643–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Van den Bossche J, and van der Windt GJW, Fatty Acid Oxidation in Macrophages and T Cells: Time for Reassessment?, Cell Metab 28 (2018), pp. 538–540. [DOI] [PubMed] [Google Scholar]
- [53].van der Windt GJ, Everts B, Chang CH, Curtis JD, Freitas TC, Amiel E, Pearce EJ, and Pearce EL, Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development, Immunity 36 (2012), pp. 68–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Divakaruni AS, Hsieh WY, Minarrieta L, Duong TN, Kim KKO, Desousa BR, Andreyev AY, Bowman CE, Caradonna K, Dranka BP, Ferrick DA, Liesa M, Stiles L, Rogers GW, Braas D, Ciaraldi TP, Wolfgang MJ, Sparwasser T, Berod L, Bensinger SJ, and Murphy AN, Etomoxir Inhibits Macrophage Polarization by Disrupting CoA Homeostasis, Cell Metab 28 (2018), pp. 490–503.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Samudio I, Harmancey R, Fiegl M, Kantarjian H, Konopleva M, Korchin B, Kaluarachchi K, Bornmann W, Duvvuri S, Taegtmeyer H, and Andreeff M, Pharmacologic inhibition of fatty acid oxidation sensitizes human leukemia cells to apoptosis induction, J Clin Invest 120 (2010), pp. 142–56. [DOI] [PMC free article] [PubMed] [Google Scholar]