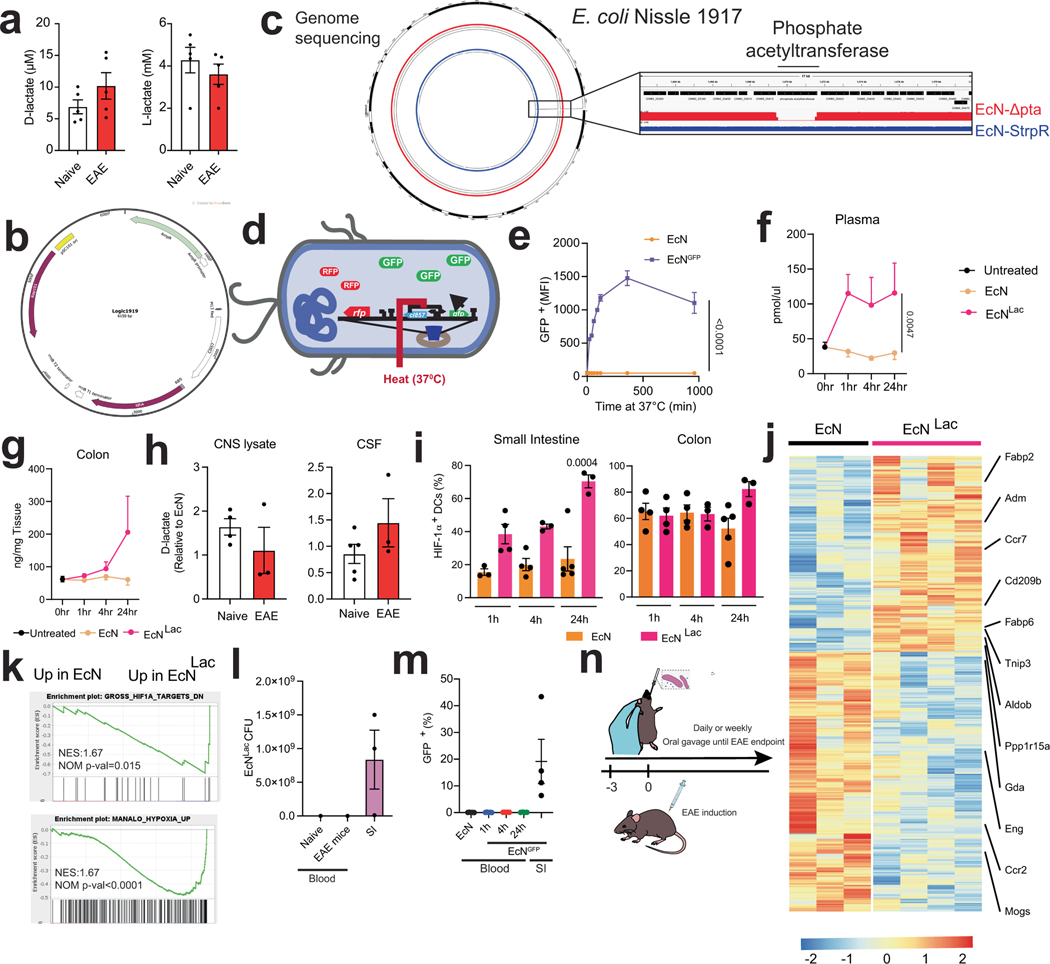
Extended Data Fig. 9 |. EcNLac characterization.
a, L-LA and D-LA concentration in plasma taken from naive and peak EAE WT mice (n = 5 per group). b, Schematic depicting the genome sequencing of EcNLac and parental EcN strains. c, Schematic for the plasmid used to induce ldhA expression in the EcNLac engineered strain. d, EcNGFP reporter strain. e, GFP expression in EcNGFP after activation at 37C. f,g, D-LA concentration in plasma (f) and colon tissue (g) after EcNLac or EcN administration (n = 5–9 mice per group). h, D-LA concentration in CSF and CNS lysate from naive (n = 4–5) and EAE (n = 3) WT mice after EcNLac administration, shown relative to D-LA concentration levels in EcN-treated mice. i, Percentage of HIF-1α+ DCs out of total DCs isolated from small intestine and colon tissue from EcN (n = 3–5) or EcNLac (n = 3–4) treated mice. j,k, Heat map ( j) and GSEA (k) analysis of RNA-seq of DCs isolated from the small intestine of EcN (n = 3) or EcNLac (n = 4) treated mice. l,m, EcNLac CFUs in blood isolated from naive and peak EAE mice treated with EcNLac daily for a week (l) and EcNGFP in blood 1, 4 and 24 h after oral gavage (m). Small intestine CFU levels are shown as positive controls (n = 4–5 per group). n, Experimental design to assess the effect of EcNLac daily or weekly administration on EAE disease course. Statistical analysis was performed using two-way ANOVA with Šídák’s post-hoc test for e,f, and one-way ANOVA with Dunnett’s post-hoc test for selected multiple comparisons for i. Data shown as mean ± s.e.m.