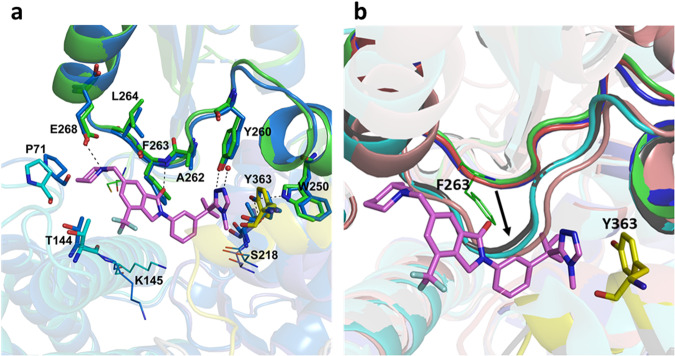
Fig. 6. Comparison of the compound binding pocket of 3mCbl-b-C7683 with that of active and inactive Cbl structures.
a An overlay of the 3mCbl-b-C7683 (green, C7683 magenta sticks) and the apo Cbl-b (blue, PDB ID: 3VGO)15 structures (both inactive). The F263-bearing loop, as well as the amino acid sidechains in the vicinity of the compound overlap well in both structures, indicating that binding of the compound does not induce changes in the binding site. b A superposition of C7683-bound, active and inactive Cbl structures. The C7683-bound structure (green), Cbl-b apo structure (blue, PDB ID: 3VGO)15 and c-Cbl apo structure (red, PDB ID: 2Y1M)16 have the F263-bearing loop in the same conformation, which represents an inactive closed Cbl conformation. The c-Cbl protein in complex with a ZAP-70 substrate peptide structure (cyan, PDB ID: 2Y1N)16, the c-Cbl protein in complex with ZAP-70 peptide and UbcH7 E2 ligase structure (gray, PDB ID: 1FBV)13, and the pTyr363 Cbl-l structure in complex with ZAP-70 peptide, UbcH5B E2 ligase, and ubiquitin (brown, PDB ID: 3ZNI)3 have the F263-bearing loop that is crucial for compound binding shifted by ~4 Å as indicated by the black arrow - which represents an open, inactive substrate-bound or active (phosphorylated) conformation.