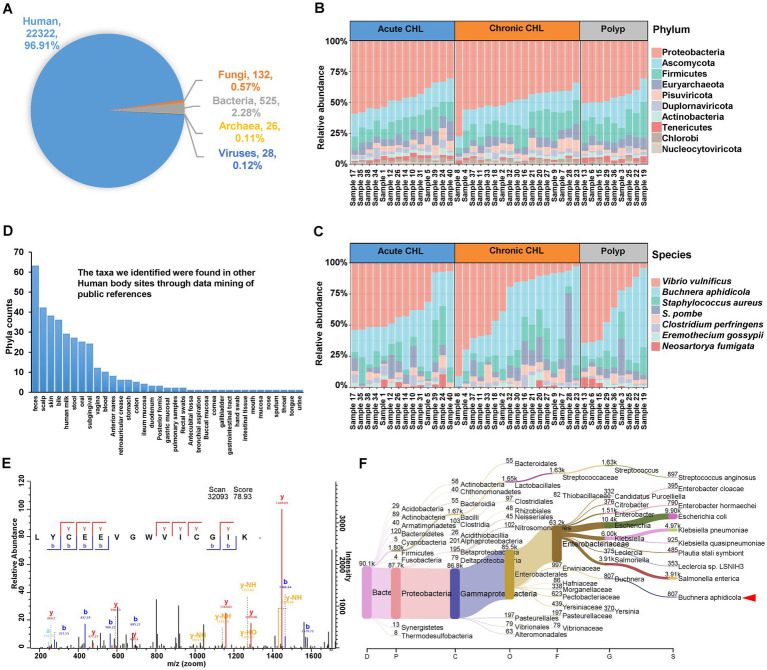
Figure 2.
Identification of microbes from gallstone and polyp bile samples. (A) The proportion of unique peptides identified in human, fungi, bacteria, archaea, and virus. (B) The structure and abundance of the top 10 phyla in both polyp and gallstone bile samples were analyzed using the intensity of identified microbial peptides. CHL, cholecystitis. (C) The structure and abundance of the top 10 microbial species in polyp and gallstone bile samples. (D) The taxa we identified were found in other Human body sites through data mining of public references. The details were deposited in Supplementary Table S2. (E) The tandem MS spectrum of peptide LYCEEVGWVICGIK identified from lepA protein (LEPA_BUCCC) of Buchnera aphidicola. (F) The Sankey plot shows the metataxonomic analysis of human bile samples. The dataset was retrieved from GenBank with the BioProject accession number PRJNA580086 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA580086). In this project, 45 biliary bile samples were analyzed using 16S rRNA sequencing. We re-analyzed the dataset against kraken2-microbial database (https://lomanlab.github.io/mockcommunity/mc_databases.html) using kraken2 program (Lu et al., 2022). The red arrowhead represents the clade of Buchnera aphidicola, which has been detected in 44 out of 45 bile samples, comprising a total of 7,313 reads.