Fig. 5. Yq12 heterochromatic region.
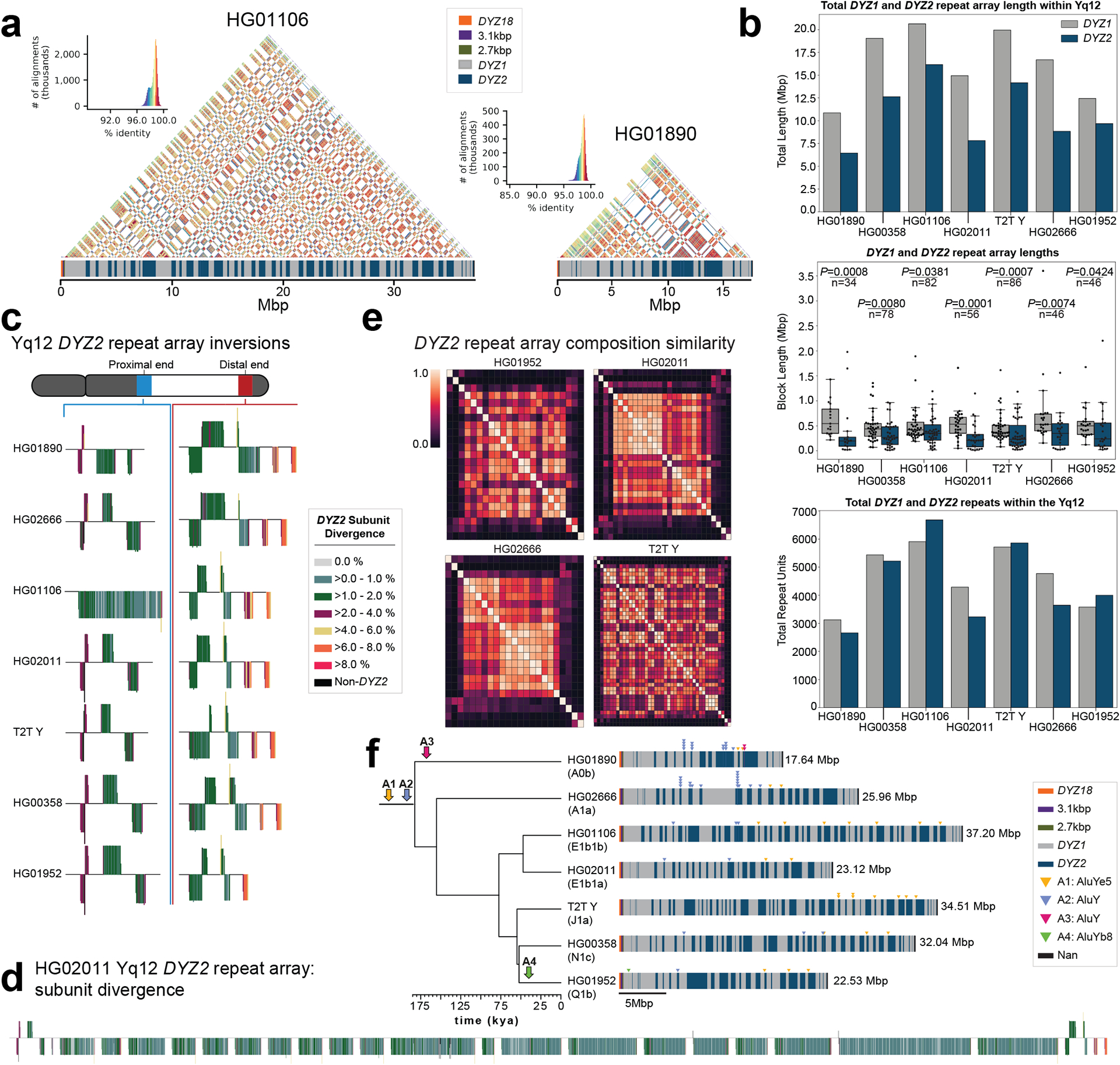
a. Yq12 heterochromatic subregion sequence identity heatmap in 5 kbp windows for two samples with repeat array annotations.
b. Bar plot of DYZ1 and DYZ2 total repeat array lengths (top), boxplots of individual array lengths (middle) and total number of DYZ1 and DYZ2 repeat units (bottom) within contiguously assembled genomes. Black dots represent individual arrays. Statistically significant p-values comparing DYZ1 and DYZ2 array lengths within each assembly and n values are shown (alpha=0.05, two-sided Mann-Whitney U test, Methods). Boxplot limits indicate quartiles, the whiskers encompass the full range of the data (except for ‘outliers’), and the median is indicated by the center line.
c. DYZ2 repeat array inversions in the proximal and distal ends of the Yq12 subregion. DYZ2 repeats are coloured based on their divergence estimate and visualized based on their orientation (sense - up, antisense - down).
d. Detailed representation of DYZ2 subunit divergence estimates for HG02011 (see panel c for colour legend).
e. Heatmaps showing the inter-DYZ2 repeat array subunit composition similarity within a sample. Similarity is calculated using the Bray-Curtis index (1 – Bray-Curtis Distance, 1.0 = the same composition). DYZ2 repeat arrays are shown in physical order from proximal to distal (from top down, and from left to right).
f. Mobile element insertions identified in the Yq12 subregion. We identified four putative Alu insertions across the seven gapless Yq12 assemblies. Their approximate location, as well as expansion and contraction dynamics of Alu insertion containing DYZ repeat units, are shown (right). Following the insertion into the DYZ repeat units, lineage-specific contractions and expansions occurred. Two Alu insertions (A1 and A2) occurred prior to the radiation of Y haplogroups (at least 180,000 years ago), while two additional Alu elements represent lineage-specific insertions. The total length of the Yq12 region is indicated on the right.
