Extended Data Fig. 3. Examples of structural variation identified in the assembled Y chromosomes.
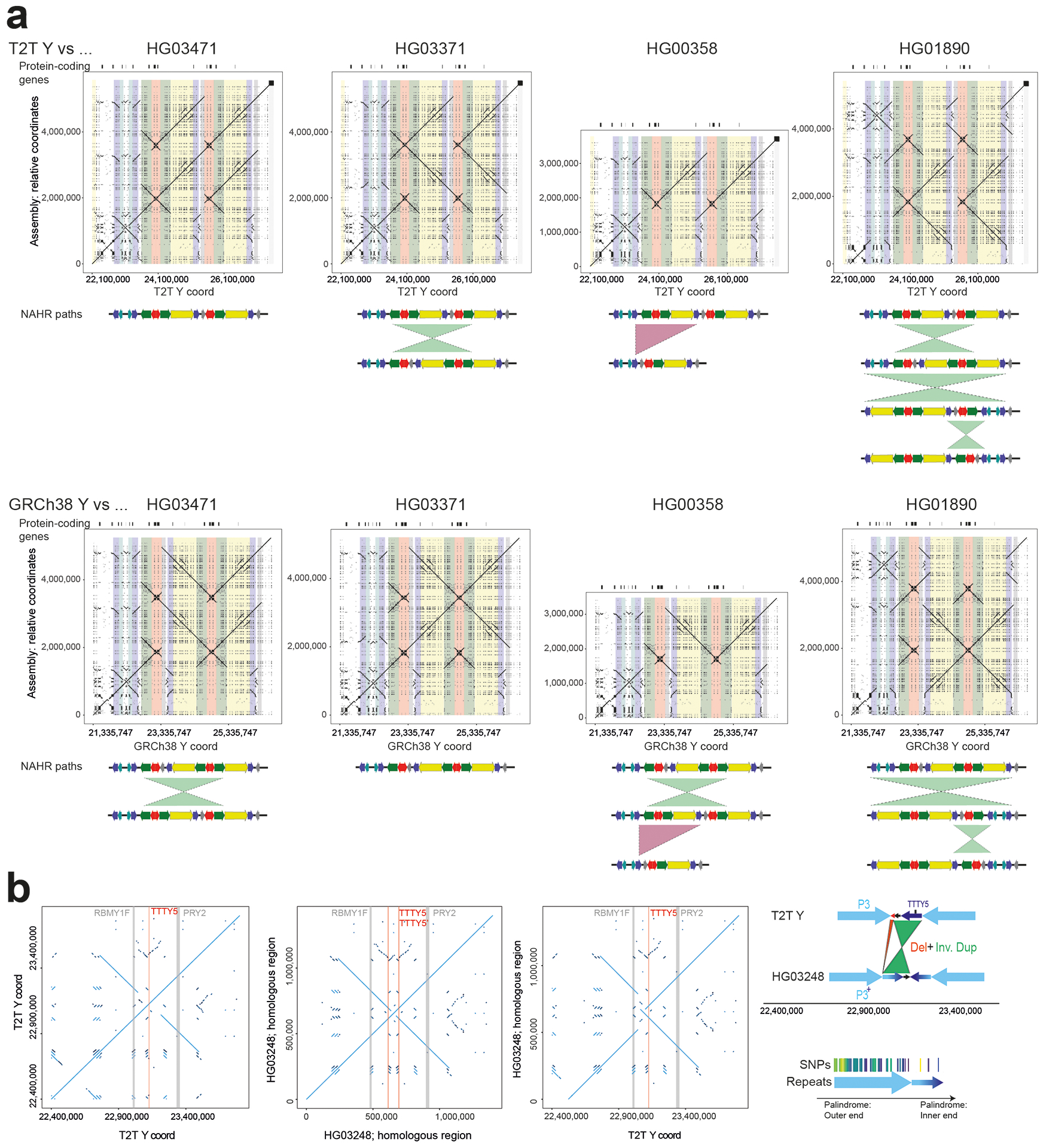
a. Inversions identified in the AZFc/ampliconic 7 subregion. Top - comparison between the T2T Y and select de novo assemblies, bottom - GRCh38 Y and the de novo assemblies (see Fig. S34 for details on AZFc/ampliconic 7 subregion composition). Potential NAHR path is shown below the dotplot.
b. Inverted duplication affecting roughly two thirds of the 161 kbp unique ‘spacer’ sequence in the P3 palindrome, spawning a second copy of the TTTY5 gene and elongating the LCRs in this region. A detailed sequence view reveals a high sequence similarity between the duplication and its template, and its placement in Y phylogeny supports emergence of this variant in the common ancestor of haplogroup E1a2 carried by NA19239, HG03248 and HG02572 (Fig. 3a).
