Figure 1.
Standardized visualization across multiple mutant landscapes using reference-based data integration
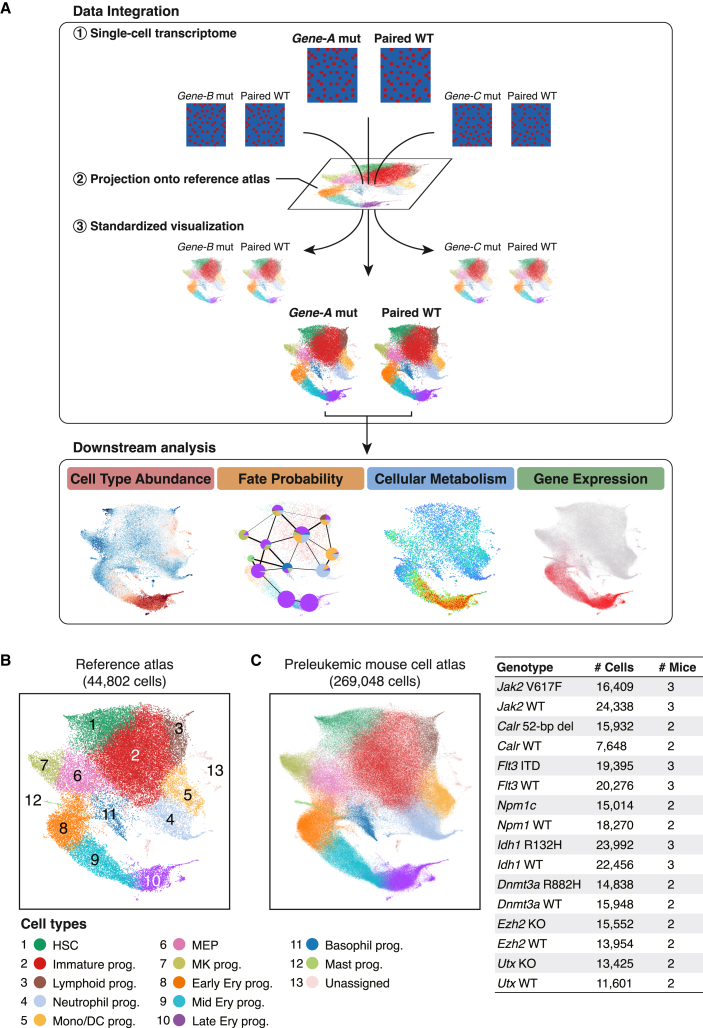
(A) Schematic overview of the analysis workflow in this study. Data integration: scRNA-seq data of individual animals are first projected onto the reference mouse hematopoietic atlas and all visual representations of the results are shown in the common UMAP space. Downstream analysis: the downstream analysis modules then infer differential abundance, cellular fate probability, cellular metabolic activity and gene expression changes. All results are shown in the common UMAP space to permit comparison among genotypes.
(B) UMAP plot of the reference mouse hematopoietic atlas11 (44,802 cells). HSC, hematopoietic stem cell; prog, progenitors; Mono, monocyte; DC, dendritic cell; MEP, megakaryocyte-erythroid progenitors; MK, megakaryocyte; Ery, erythroid.
(C) Integrated preleukemic mouse hematopoietic atlas (38 animals, 269,048 cells). Cells are color coded according to cell types as in (B). WT, wild type; KO, knockout.