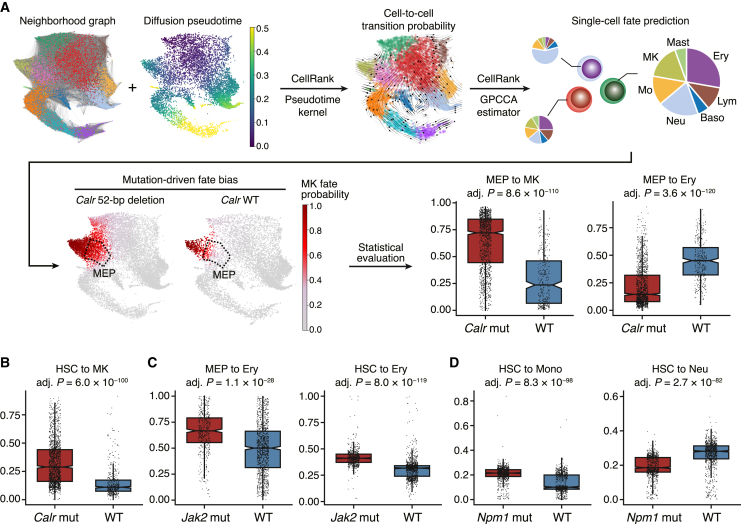
Figure 3.
Mutation-driven fate bias in early HSPCs
(A) Workflow of CellRank-based differential fate probability analysis. A neighborhood graph and diffusion pseudotime were computed (top left) and used to infer cell-to-cell transition probabilities (top middle). On the basis of the transition matrix, the fate probabilities toward seven hematopoietic lineages (megakaryocyte, erythroid, lymphoid, neutrophil, monocyte, basophil, and mast cell) were estimated for the individual cells (top right). The single-cell estimates of fate probabilities were then compared between the paired mutant and wild-type samples (bottom). Ery, erythroid; Lym, lymphoid; Baso, basophil; Neu, neutrophil; Mo, monocyte; MK, megakaryocyte.
(B) Significant difference in the megakaryocyte probability between Calr mutant and wild-type HSCs.
(C) Significant differences in the erythroid probability between Jak2 mutant and wild-type MEPs (left) and HSCs (right).
(D) Significant differences in the monocyte (left) and neutrophil (right) probability between Npm1 mutant and wild-type HSCs. Boxplots show median and first/third quartiles. The whisker extends from the smallest to the largest values within 1.5 × IQR from the box hinges. p values are from logistic regression and likelihood ratio test and are BH adjusted.