Figure 5.
Perturbed regulation of gene expression in preleukemic mutant models
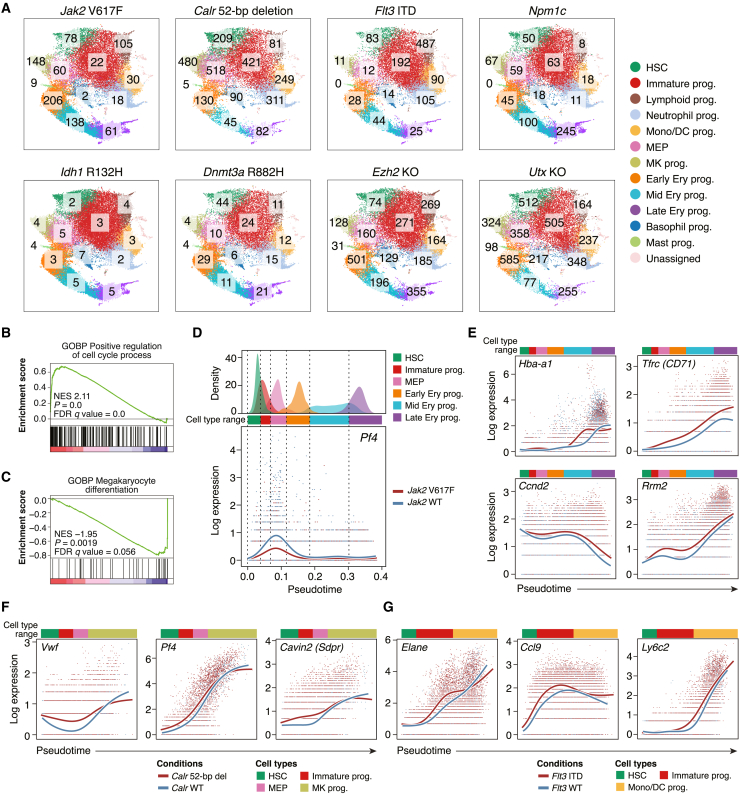
(A) The number of genes differentially expressed in each cell type from each mutant model compared with the wild-type counterpart. The numbers are shown over the corresponding cell types in the individual UMAP plots. HSC, hematopoietic stem cell; prog, progenitors; Mono, monocyte; DC, dendritic cell; MEP, megakaryocyte-erythroid progenitors; MK, megakaryocyte; Ery, erythroid.
(B) Significant upregulation of cell cycle regulators in Jak2 mutant early erythroid progenitors. Gene Ontology (GO) terms for biological processes (BPs) were evaluated. NES, normalized enrichment score; FDR, false discovery rate.
(C) Significant downregulation of genes regulating megakaryocytic differentiation in Jak2 mutant MEPs.
(D) Differential expression dynamics of Pf4 between the erythroid trajectory of the Jak2 mutant and the paired wild-type samples. The upper panel shows the pseudotime distribution of each cell type, defining the pseudotime ranges of dominant cell types. The lower panel shows the pseudotemporal expression patterns of Pf4. The red (Jak2 V617F) and blue (wild-type) lines show the expression smoothers estimated by a negative binomial generalized additive model.41 Each dot shows the log-normalized expression and the pseudotime of each cell.
(E–G) Significantly altered gene expression patterns in the Jak2 mutant and wild-type erythroid trajectory (E), the Calr mutant and wild-type megakaryocyte trajectory (F), and the Flt3 mutant and wild-type myelomonocytic trajectory (G). The pseudotime ranges of dominant cell types are indicated with colored bars.