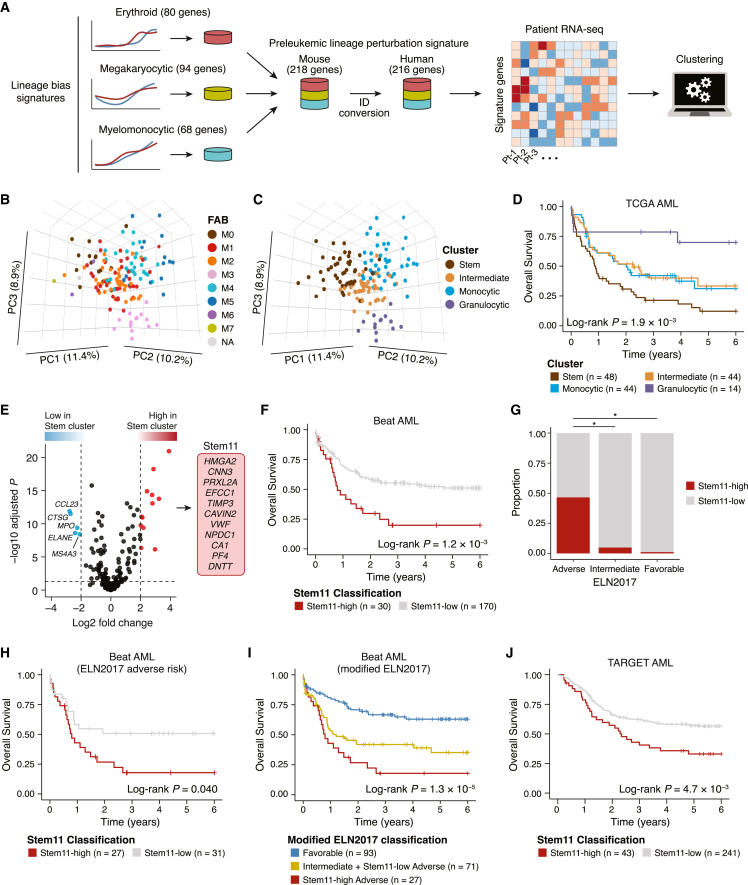
Figure 6.
Prognostic relevance of preleukemic lineage perturbation signature genes
(A) Overview of the analysis of patient RNA-seq data. The erythroid, megakaryocytic, and myelomonocytic bias signatures derived from the pseudotemporal differential expression analysis were combined to develop the PLPS (preleukemic lineage perturbation signature) genes, which were then used to cluster the patient data.
(B and C) Principal-component analysis of the TCGA cohort. Samples are colored according to the FAB classification (B) and the patient clusters (C). PC1, PC2, and PC3 represent the first three principal components, with the percentage of variance explained indicated on the axes.
(D) Survival analysis comparing the overall survival of the different clusters of TCGA AML cohort.
(E) Volcano plot showing the differential expression of PLPS genes. Genes with significant upregulation (BH-adjusted p < 0.05 and log2 fold change > 2; red) and downregulation (BH-adjusted p < 0.05 and log2 fold change < −2; blue) are color coded.
(F) Survival analysis comparing the Stem11-high and Stem11-low groups in the Beat AML cohort.
(G) Proportions of Stem11-high patients in each of the ELN2017 risk groups. p values are from Fisher’s exact test. ∗p < 1.0 × 10−5.
(H) Survival analysis comparing the Stem11-high and Stem11-low patients among the ELN2017 adverse risk group in the Beat AML cohort.
(I) Survival analysis comparing our modified ELN2017 risk groups, where the Stem11-low, ELN2017 adverse-risk patients were re-stratified to the intermediate risk group.
(J) Survival analysis comparing the Stem11-high and Stem11-low groups in the TARGET AML cohort.