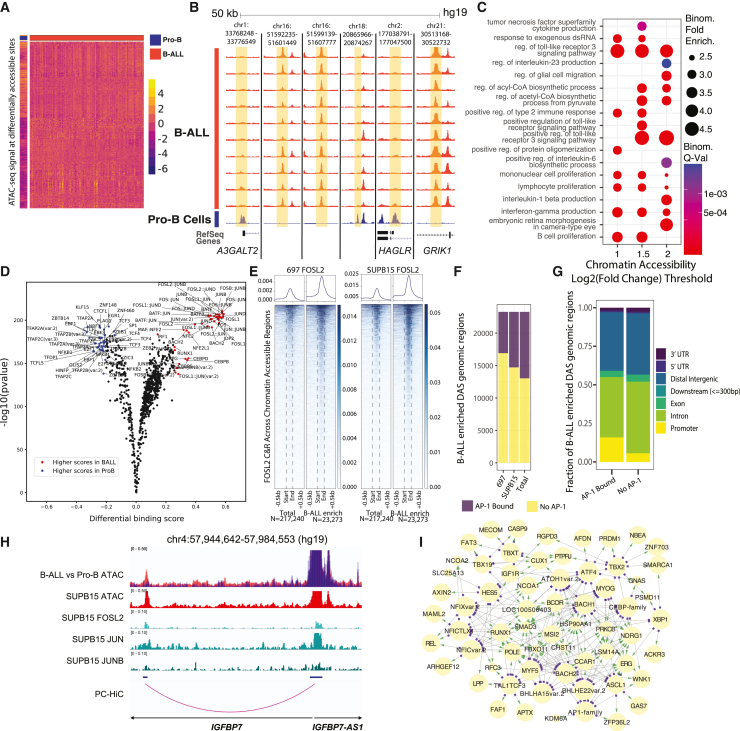
Figure 3.
Mapping differential accessibility between B-ALL and Pro-B cells
(A) Heatmap using row-wise hierarchical clustering of Pro-B cell or B-ALL patient sample variance-stabilized ATAC-seq signal as Z score across Pro-B cell and B-ALL-enriched DASs. DASs within heatmap are >1 or <−1 log2-adjusted fold change.
(B) ATAC-seq signal-track examples of top Pro-B-cell-enriched DASs and B-ALL-enriched DASs on the UCSC genome browser. Flanking genomic regions are included for context.
(C) Gene ontology analysis of DASs with higher accessibility in B-ALL (B-ALL enriched) at various log2-adjusted fold-change thresholds. All terms were significant using both binomial and hypergeometric statistical tests.
(D) Differential TF footprinting between Pro-B cells and B-ALL patient samples across 217,240 B-ALL genomic regions of interest.
(E) FOSL2 CUT&RUN enrichment heatmaps at all B-ALL accessible chromatin sites (on left, N = 217,240 regions) and B-ALL-enriched DASs (on right, N = 23,273 regions) in SUPB15 (left enrichment heatmap) and 697 (right enrichment heatmap) cells. Total numbers of sites are shown below each heatmap. Rows in adjacent pairs of heatmaps are unaligned.
(F) Number of B-ALL-enriched DASs overlapping AP-1 TF occupancy (FOSL2, JUN, and/or JUNB) in 697 (left), SUPB15 (middle), and both B-ALL cell lines (right). Numbers of overlapping sites are shown in purple while non-overlapping sites are shown in yellow.
(G) Genome annotation of B-ALL-enriched DASs with AP-1 TF occupancy (left) or that are devoid of AP-1 TF occupancy (right).
(H) IGV genome browser image showing a B-ALL-enriched DAS that maps to accessible chromatin and sites of AP-1 TF occupancy in SUPB15 cells. Promoter capture Hi-C (PC-HiC) looping between the distal AP-1 occupied sites and the IGFBP7 gene promoter is shown. B-ALL (red) and Pro-B (blue) cell ATAC-seq tracks are overlaid in the top panel. Signal tracks for FOSL2, JUN, and JUNB in SUBP15 cells are shown.
(I) TF and target gene network of DASs with higher accessibility in B-ALL (B-ALL-enriched). Network is subset for top TF footprints across DASs ranked by the top mean log2-adjusted fold-change transcription factor footprint signal. Target genes determined with B-ALL patient origin promoter capture Hi-C are subset for a cancer-implicated gene set ranked by the top expressed genes. Network connections are colored as TFs (purple blocks) to target gene (green arrowheads) pairs. Select expansive and highly similar TF motif families are grouped (AP-1 and CEBP, AP1 family and CEBP family).