FIGURE 1.
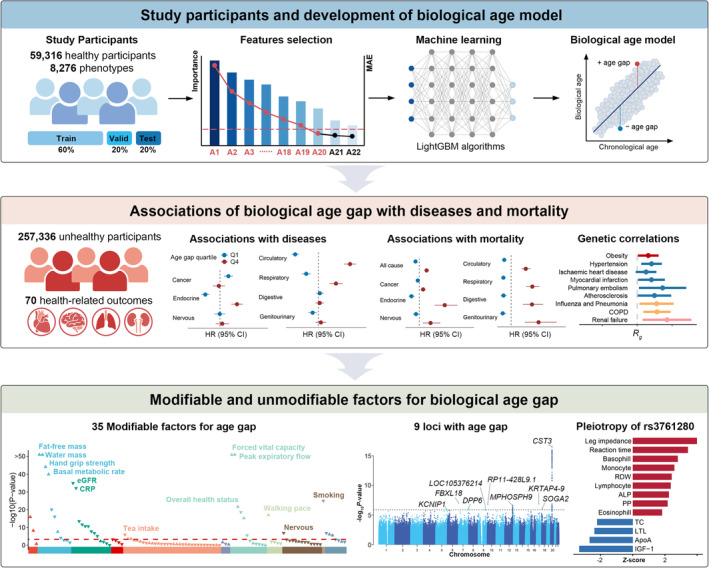
Graphical abstract of the study. Top part, the study participants and development of biological age model. The study included 59,316 healthy participants in the UK Biobank and considered 8276 phenotypes for developing biological age model. All healthy participants were further divided into training set (60%), validation set (20%), and testing set (20%). LightGBM algorithm was conducted to identify the most important predictors for biological age and build the model and the top 20 predictors were selected. Then the age gap, the difference between the estimated biological age and chronological age, was calculated within the participants. Middle part, the associations of age gap with diseases and mortality. We tested the longitudinal associations of age gap with 70 common health‐related outcomes, all‐cause mortality and cause‐specific mortality, and the genetic correlations of age gap with common health‐related outcomes. Bottom part, the modifiable and unmodifiable factors for age gap. We identified 34 modifiable factors and 9 genomic risk loci for age gap and profiled the pleiotropy of rs3761280 in the UK Biobank. ALP, alkaline phosphatase; ApoA, apolipoprotein A; CI, confidence interval; COPD, chronic obstructive pulmonary disease; CRP, C‐reactive protein; eGFR, estimated glomerular filtration rate; HR, hazard ratio; IGF‐1, insulin growth factor 1; LightGBM, Light Gradient Boosting Machine; LTL, leukocyte telomere length; PP, pulse pressure; RDW, red blood cell distribution width; TC, total cholesterol.
