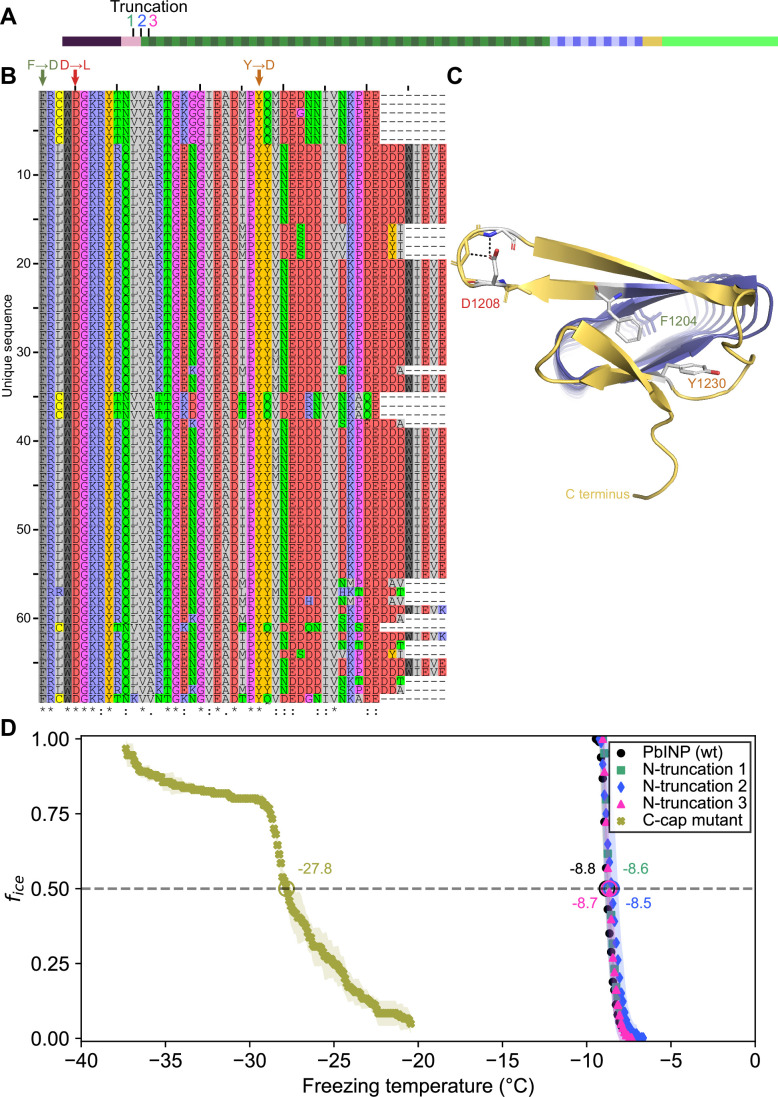
Figure 7. Investigating putative INP cap structures through a series of mutant proteins.
(A) Sites of N-terminal truncations to PbINP, indicating the location of the starting residue in the shortened construct. (B) Alignment of representative ice nucleation protein (INP) C-terminal domains from the genus Pseudomonas. Mutated residues and their one-letter codes are indicated above. Symbols at the bottom indicate consensus (* for fully conserved, : for conservation of strongly similar chemical properties, . for conservation of weakly similar chemical properties). (C) Predicted location of mutated residues in the PbINP C-terminal cap with side chains shown and predicted H-bonds for D1208 shown as dashed lines. (D) The ice nucleation curves for the N- and C-terminal cap mutants.