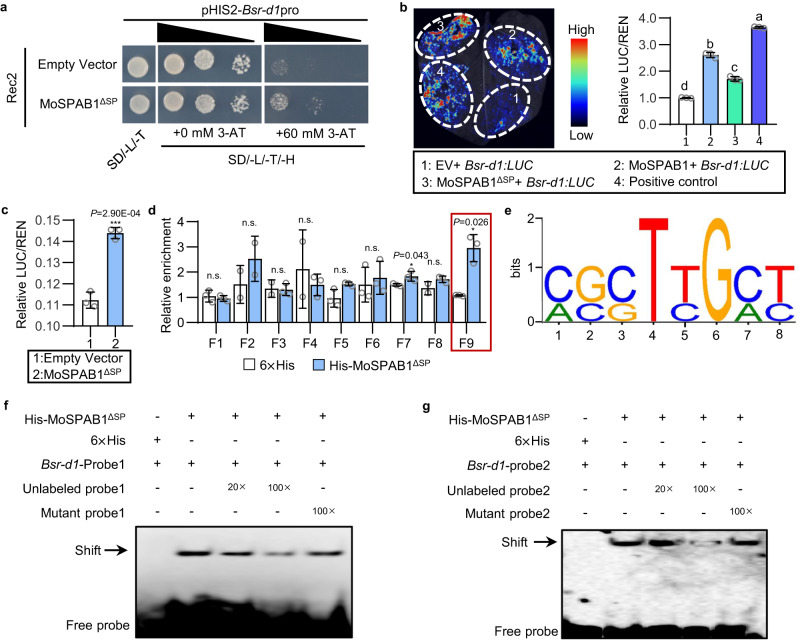
Fig. 1. M. oryzae MoSPAB1 directly binds to the Bsr-d1 promoter.
a Binding of MoSPAB1ΔSP to the Bsr-d1 promoter in yeast one-hybrid assay. b Activation of the Bsr-d1 promoter by MoSPAB1 in luciferase assay using N. benthamiana. Luciferase signals were imaged (left panel) and measured (right panel) using a dual-LUC assay 48 hr after infiltration. The positive control contains transcription factor EAT1 and OsLTPL94 promoter driving reporter. EV contains 35 S:YFP as the negative control. firefly luciferase (LUC) activities were normalized to Renilla luciferase (REN). Values are mean ± SD, n = 3 biologically independent samples. Data analyzed by one-way ANOVA followed by two-sided least significant difference (LSD) test for multiple comparisons. Different letters indicate significant differences (P < 0.05). P-values are shown in the Source Data file. c Luciferase activity assay for MoSPAB1ΔSP binding to the Bsr-d1 promoter in rice protoplasts. The fluorescence signals of LUC and REN (internal control) were detected after rice protoplasts were co-transfected with the vectors for 16 h. Values are mean ± SD, n = 3 biologically independent samples (Student’s two-sided t-test, ***P < 0.001). d DNA affinity purification (DAP)-qPCR analysis for enrichment of Bsr-d1 promoter DNA by recombinant His-MoSPAB1ΔSP protein. The ratio of His-MoSPAB1ΔSP/6×His ≥ 2 was used as the threshold for significance. Values are mean ± SD, n = 3 biologically independent samples (Student’s two-sided t-test, *P < 0.05, n.s. indicates no significance). e A conserved sequence bound by MoSPAB1ΔSP was identified using the MEME suite version 5.4.1. f Binding of His-MoSPAB1ΔSP to probe 1 of the Bsr-d1 promoter. Similar results are obtained from three independent biological experiments. g Binding of His-MoSPAB1ΔSP to probe 2 of the Bsr-d1 promoter. Similar results are obtained from three independent biological experiments. Source data are provided as a Source Data file.