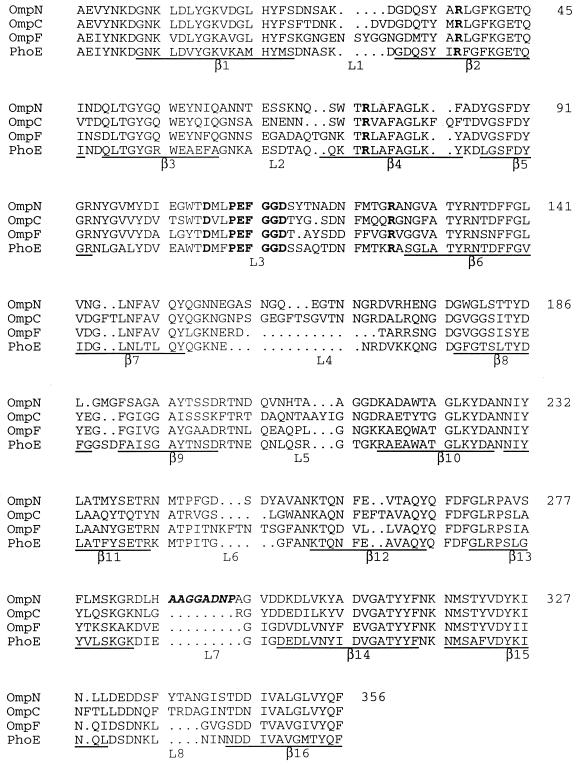
FIG. 2.
Comparison of the predicted amino acid sequence of OmpN with sequences of various known E. coli porins. Sequences underlined below each block correspond to the β strands in the three-dimensional structures of OmpF and PhoE (6). A highly conserved porin-specific sequence motif, PEFGGD (14), in loop L3, and five conserved charged residues (R37, R75, D106, E110, and R126) which form a strong transversal electrostatic field in the channel interior are shown in bold. The additional amino acid residues, present in the predicted surface-exposed loop L7 in OmpN, are shown in bold italics. The overall similarity of OmpN is highest to OmpC (65% identical residues), followed by PhoE (62% identical residues) and OmpF (58% identical residues).