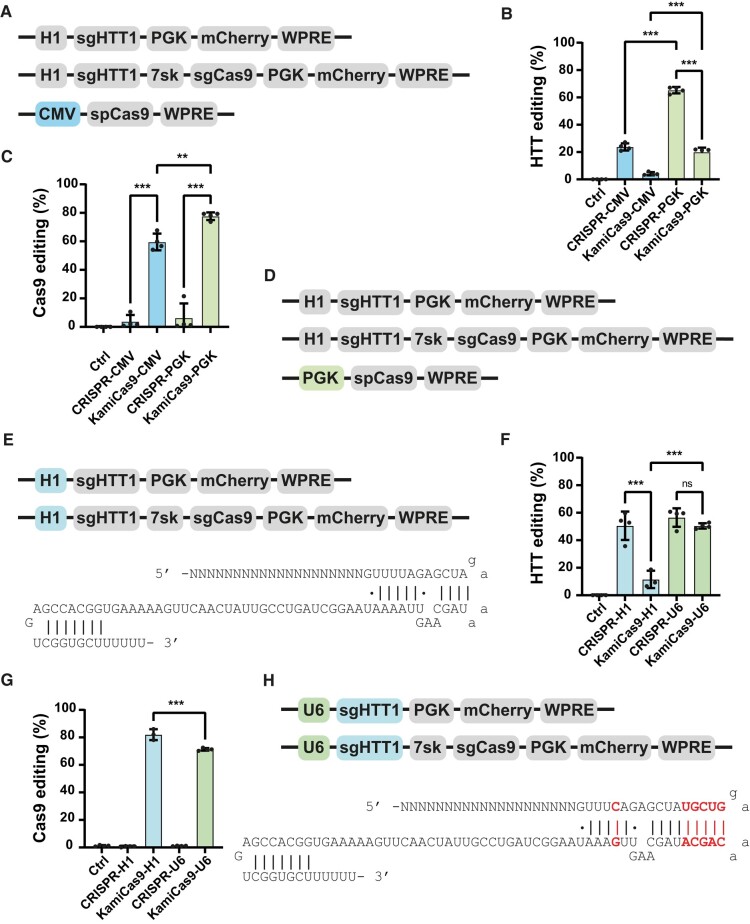
Figure 1.
Optimization of the LV-KamiCas9 system in human neuronal progenitor cells (NPCs). (A) Schematic representation of the original constitutive and KamiCas9 vectors used for in vitro gene editing. The first LV expresses the sgRNA targeting the human HTT (sgHTT1) under the control of the H1 polymerase III promoter and mCherry under the control of the mouse phosphoglycerate kinase I promoter (PGK). The second vector is the KamiCas9 version with an additional cassette expressing a sgRNA targeting the translation start site of spCas9 under the control of the 7sk promoter. The third plasmid encodes the spCas9 nuclease under the control of a cytomegalovirus (CMV) promoter. (B) Human HTT editing was assessed three weeks after the transduction of NPCs. DNA extracts were used for TIDE analysis (n = 4 samples/group) and to compare the performances of the constitutive clustered regularly interspaced short palindromic repeats (CRISPR) and KamiCas9 systems. (C) The frequency of indels in the SpCas9 transgene was assessed in the same samples (n = 4 samples/group). (D) Schematic representation of the second-generation LV-KamiCas9 system. In this version, the only modification is the replacement of the CMV promoter with a PGK promoter to drive expression of the spCas9 nuclease gene. (E) Schematic representation of the first-generation LV-KamiCas9 system. In this version, the expression of the sgRNA targeting HTT is under the control of the H1 promoter and the original tracrRNA. (F and G) We assessed human HTT and spCas9 editing with the optimized system in NPCs (n = 4 samples/group). (H) Schematic representation of the final optimized LV-KamiCas9 system. In this optimized version, the expression of the sgRNA targeting HTT is under the control of the U6 promoter and an optimized tracrRNA. Results are presented as the mean ± SD. Statistics: one-way ANOVA and multiple comparisons with Tukey correction: ****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05.