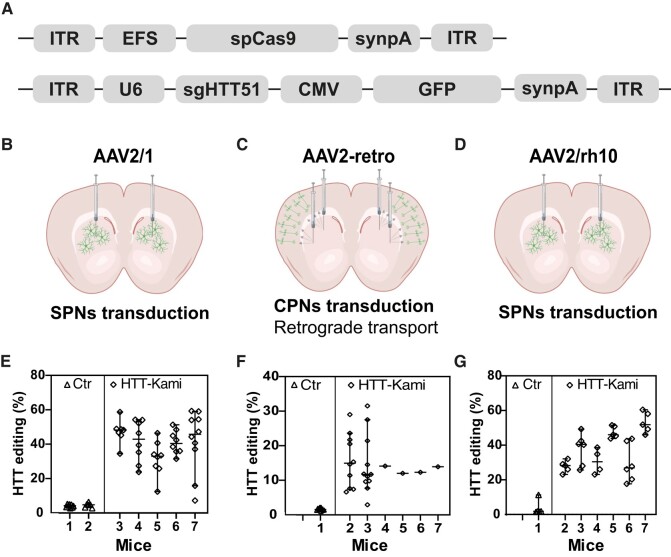
Figure 3.
Htt editing in wild-type mice. (A) Schematic representation of the AAV vectors used for in vitro gene editing. The first plasmid encodes the Streptococcus pyogenes Cas9 (spCas9) nuclease under the control of the short elongation factor (EFS) promoter in an AAV vector containing inverted terminal repeats (ITR) and a synthetic polyadenylation site (synpA). The second vector encodes sgHTT51 under the control of the U6 promoter and green fluorescent protein (GFP) under the control of the CMV promoter. SynpA, synthetic polyadenylation signal. (B) AAV2 serotype 1 (AAV2/1) (n = 2 Ctr, n = 5 treated mice), (C) AAV2 variant with retrograde properties (AAV2.retro) (n = 2 Ctr, n = 6 treated mice) and (D) AAV2 serotype 10 (AAV2/rh10) (n = 1 Ctr, n = 6 treated mice) were used to transduce spiny projection neurons (SPN) and/or cortical projection neurons (CPN), making use of the retrograde transport properties of AAV2.retro (schemes created with Biorender.com). (E) Htt editing in the striatum of mice treated with AAV2/1 (n = 6–8 punches/animal). Statistics: nested two-tailed t-test: P = 0.0004, t(8.477), df(5) and median with 95% CI are indicated. (F) Htt editing in the cortex of animals treated with AAV2.retro (n = 10 punches/mouse for three animals; and pooled punches/mouse for four animals). Statistics: nested two-tailed t-test: P < 0.0001, t(5.553), df(33)) and median with 95% CI are indicated. (G) HTT editing in the cortex of animals treated with AAV2/rh10 (n = 5 punches/animal) Statistics: nested two-tailed t-test: P = 0.0294, t(3.02), df(5)) and median with 95% CI are indicated.