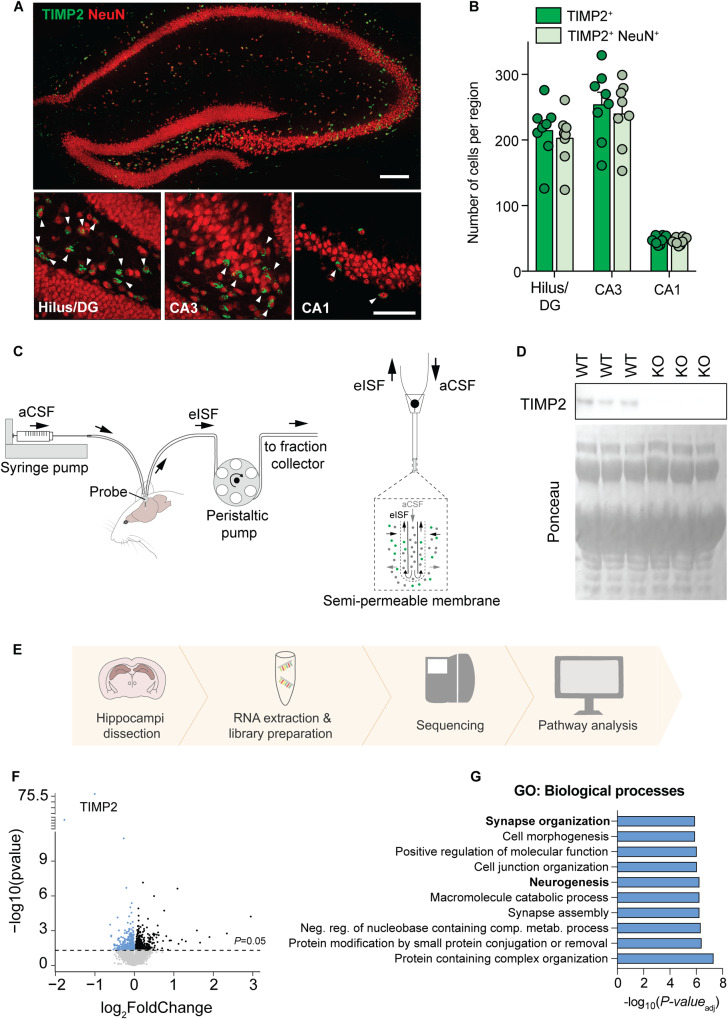
Fig. 1. TIMP2 is expressed in neurons of the hippocampus, and its deletion induces transcriptomic changes.
A Low-magnification view (upper image) of mouse hippocampus and high-magnification view (lower images) of hilus/DG, CA3, and CA1 subregions showing TIMP2+ cells and TIMP2+ cells staining for pan-neuronal nuclei marker NeuN. Scale bars, 200 μm and 20 μm (inset). B Quantification of the total number of TIMP2+ cells and TIMP2+ NeuN+ cells across hippocampal subregions in WT mice (2 months of age; N = 8, males and females). C Schematic representation of high molecular-weight cut-off (1-MDa) in vivo microdialysis for assessing TIMP2 levels in mouse hippocampal ISF. D TIMP2 immunoblotting of hippocampal ISF dialyzed from 2-month-old WT and TIMP2 KO mice, with corresponding Ponceau S stain. E Schematic representation of bulk RNA-seq workflow performed in isolated WT and TIMP2 KO hippocampi (N = 13–17 mice/group, sex-matched) for transcriptomic analysis. F Volcano plot showing the fold-change of genes (log2 scale) differentially expressed in hippocampus of TIMP2 KO vs. WT mice. Downregulated DEGs at nominal P < 0.05 are highlighted in blue (upregulated in black). G Top 10 significant pathways for downregulated DEGs from Gene Set Enrichment Analysis. Data are represented as mean ± SEM. DG dentate gyrus, eISF exchangeable interstitial fluid, aCSF artificial cerebrospinal fluid.