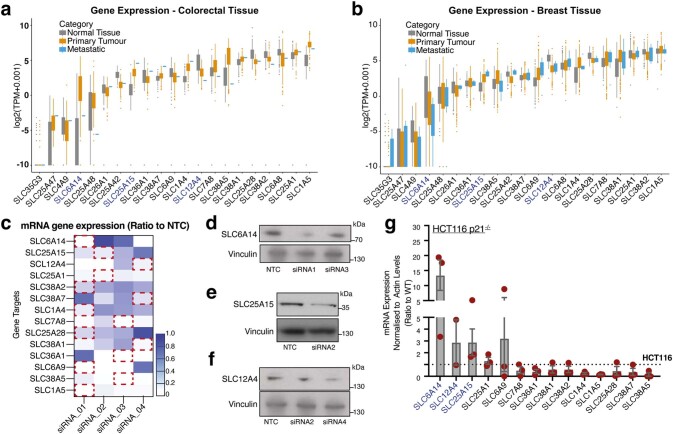
Extended Data Fig. 1. Phenotypic landscape of SLC superfamily reveals SLCs influencing serine transport in cancer cells.
a. Gene expression levels of selected SLC-encoding genes in human colorectal tissues. Values are log2(TPM + 0.001) from n = 359 normal tissue, n = 380 primary tumor and n = 1 metastatic samples. b. Gene expression levels of selected SLC-encoding genes in human breast tissues. Values are log2(TPM + 0.001) from n = 292 normal tissue, n = 1092 primary tumor and n = 7 metastatic samples. c. Heat map showing gene silencing efficiency of 4 deconvoluted siRNAs against each SLC-encoding gene. Values are mRNA expression normalized to ACTIN levels and relative to NTC control. Red boxes indicate selected siRNA for further experiments. d. Immunoblots of SLC6A14 and Vinculin (loading control) in control (NTC) or SLC6A14-silenced HCT116 cells. Blots are representative of three independent experiments. e. Immunoblots of SLC25A15 and Vinculin (loading control) in control (NTC) or SLC25A15-silenced HCT116 cells. Blots are representative of three independent experiments. f. Immunoblots of SLC12A4 and Vinculin (loading control) in control (NTC) or SLC12A4-silenced HCT116 cells. Blots are representative of three independent experiments. g. mRNA expression levels of indicated SLC-encoding genes in HCT116 p21-/- cells versus HCT116 WT control. Values are mean±s.e.m. from n = 3 independent experiments. Data in a,b:: Box and whiskers plots are log2 (TPM + 0.001) with box limits representing 25th and 75th percentiles of the dataset. Whiskers extend from the hinge to the largest or smallest value, corresponding to the upper or lower 1.5x IQR (inter-quartile range). Individual dot points represent data beyond the end of the whiskers.