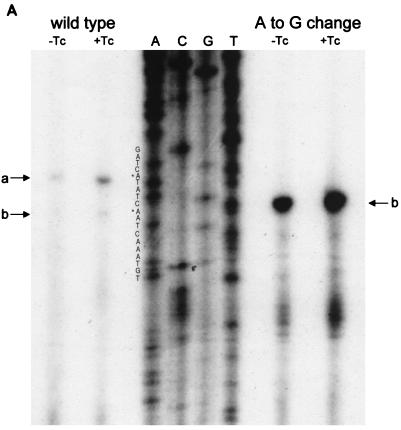
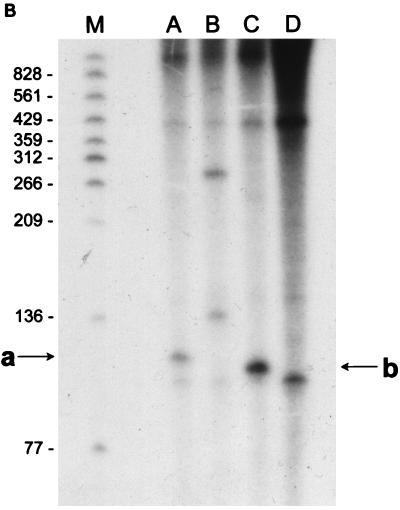
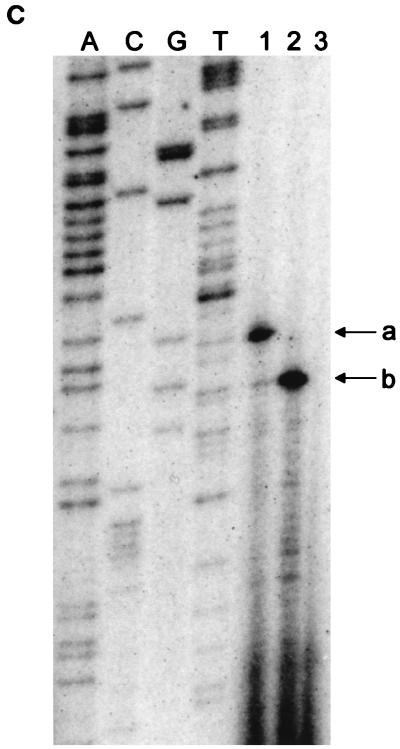
FIG. 3.
(A) Reverse transcriptase analysis of in vivo tetA(L) mRNA. RNA was isolated from the wild-type strain and from the strain containing the A→G change in the tetA(L) promoter. Strains were grown in the absence or presence of 0.25 μg of TET/ml. Transcriptional start sites are marked a and b, as in Fig. 1. Lanes A, C, G, and T are a sequence ladder obtained with the primer used for the reverse transcriptase reactions. The complement of the ladder sequence, which is the same as the sequence shown in Fig. 1C, is shown on the left of the sequencing ladder. Transcriptional start sites are marked by asterisks. The amount of radioactivity in the bands was quantitated with a PhosphorImager (Molecular Dynamics). (B) In vitro transcription. Plasmid pAN583 contained either a wild-type tetA(L) promoter-leader DNA fragment (lanes A and B), a promoter-leader DNA fragment with the A→G change (lane C), or no tetA(L) DNA (lane D). Plasmid DNA was linearized with XbaI (lanes A, C, and D) or with HindIII (lane B). Digestion with HindIII linearized the plasmid at a site 25 bp downstream of the XbaI site. Comparison of lanes A and B demonstrates that the band indicated by “a” at the left represents transcription from transcriptional start site a. In lane C, the transcription product migrates faster and represents transcription from transcriptional start site b. A nonspecific transcription product runs below band b and is present in all lanes, including lane D [no tetA(L) DNA]. Lane M contained end-labeled DNA fragments from a TaqI restriction endonuclease digest of pSE420 (2). Sizes (in base pairs) of the labeled DNA fragments are indicated on the left. (C) Reverse transcriptase analysis of in vitro-transcribed tetA(L) RNA. Lane 1, wild-type tetA(L) RNA; lane 2, mutant (A→G change) tetA(L) mRNA; lane 3, no RNA (labeled primer only). Transcriptional start sites are marked a and b, as in Fig. 1. Lanes A, C, G, and T are a sequence ladder obtained with the primer used for the reverse transcriptase reactions.