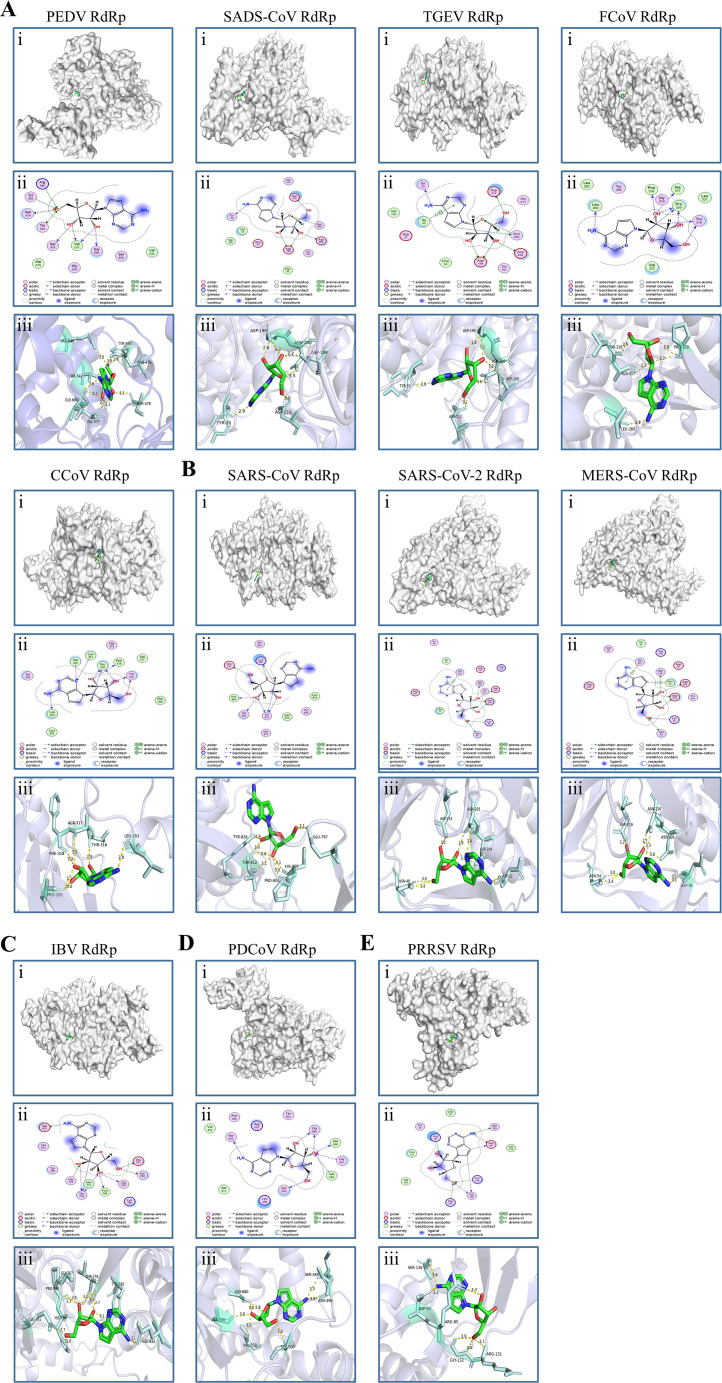
Fig. 9.
Molecular docking of Tubercidin with RdRp of nidoviruses. Tubercidin was docked with RdRp proteins from viruses including (A) α-coronaviruses (PEDV, SADS-CoV, TGEV, FCoV and CCoV), (B) β-coronaviruses (SARS-CoV, SARS-CoV-2, and MERS-CoV), (C) γ-coronavirus (IBV), (D) δ-coronavirus (PDCoV) and (E) Arteritis virus (PRRSV) using Autodock. (i) Cartoon representation, overlay of the crystal structures of Tubercidin and viruses RdRp protein were illustrated. (ii) 2D interactions of Tubercidin and viruses RdRp protein. (iii) Three-dimensional structures of the binding pockets were showed by PyMOL software. The proteins and compound are represented as cartoons and sticks, respectively. The RdRp proteins' three-dimensional structures are colored light white. The Tubercidin's two-dimensional structure is colored green, red, and blue. The Pymol software displays the sticks structure (Pale cyan) of amino acids residues in the virus RdRp protein. These amino acids residues are connected through H-bonds (yellow dotted lines) to Tubercidin, which is stabilized in the active pocket. The binding energy of the Tubercidin–viruses RdRp protein, calculated using Autodock, is listed (Table 1).