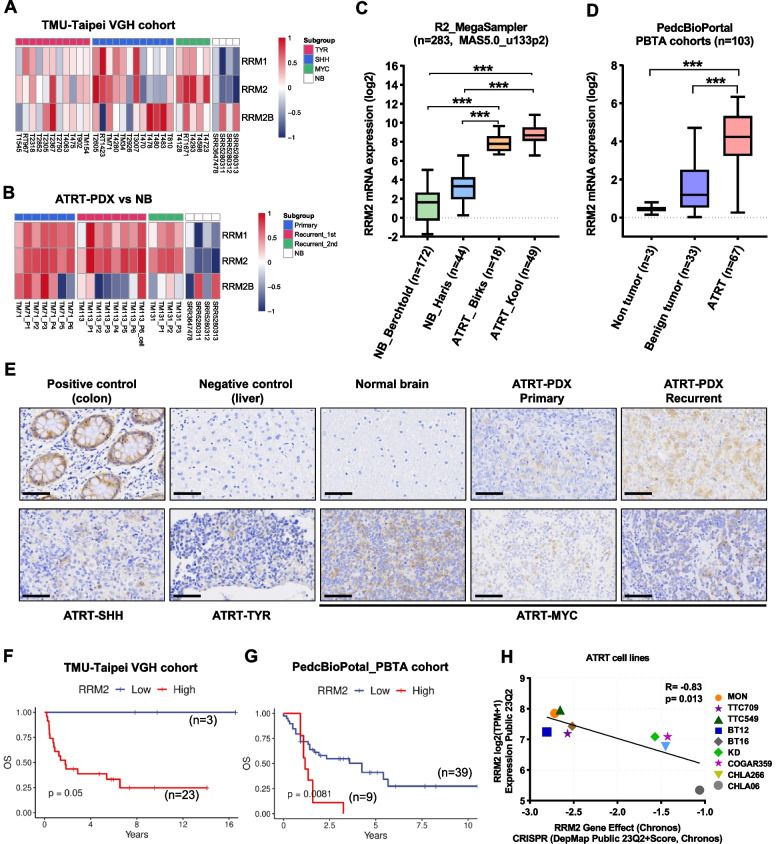
Fig. 1.
RRM2 expression and its correlation with overall survival in ATRT. A, B Heatmaps of RRM2-related gene expression in human ATRT (A) and ATRT-PDX (B) samples compared with normal brain tissues. For normal samples, raw RNA-seq data of four normal brain tissues were downloaded from the GEO data set. C Expression levels of RRM2 mRNA in human ATRT and normal brain tissues in public data sets using the R2 Platform. D Expression levels of RRM2 mRNA in human ATRT, normal brain, and benign tumor tissues in PedcBioPortal dataset. Bar indicates the mean mRNA levels of each group, data are presented as min to max, Tukey’s multiple comparisons test, ***p < 0.001. RNA expression was normalized using logarithm base 2. E Comparison of IHC staining for RRM2 in the colon (as positive control), liver (as negative control), normal brain, human ATRT, and ATRT-PDX samples. Scale bar 30 µm. F, G The correlation of RRM2 mRNA level with patient’s overall survival (OS) in TMU-Taipei VGH cohort (n = 26) (F), and PBTA cohort from PedcBioPortal (n = 48) (G). H The correlation between RRM2 gene effect and RRM2 gene expression in nine ATRT cell lines was analyzed from DepMap project. Gene effect means the necessity of RRM2 for the survival of ATRT cell lines was retrieved and analyzed from CRISPR (DepMap Public 23Q2 + Score, Chronos) dataset. A lower score means that a gene is more likely to be dependent in a given cell line, meaning that gene has a higher contribution to cell survival. The RRM2 expression was analyzed from the project Expression Public 23Q2 dataset. Gene expression TPM values of the protein-coding genes for DepMap cell lines. Values are inferred from RNA-seq data using the RSEM tool and are reported after log2 transformation, using a pseudo-count of 1; log2(TPM + 1)