Figure 1:
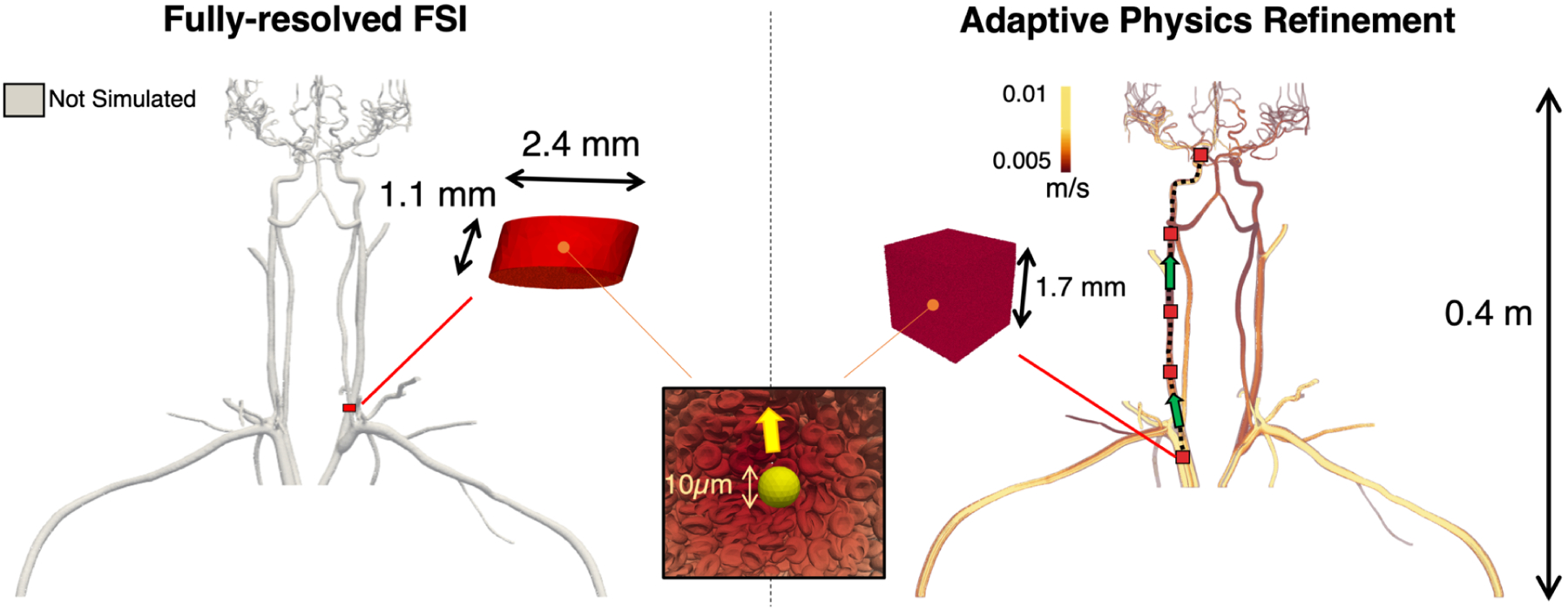
Use of the Adaptive Physics Refinement (APR) model within an upper body vasculature enables up to 4 orders of magnitude increase in total simulated fluid volume accessible to cellular resolution. Using 256 nodes on Summit (1.5k GPUs and 10.8k CPUs), the APR simulation, shown on the right, has the ability to track a cancer cell through the upper body geometry, a volume of 41.0 mL, using a local finely resolved window. This window can travel through the vessel, depicted by the red boxes moving along the dashed black line in the direction of the green arrows, opening up the entire volume to a submicron, cell-resolved mesh. By comparison, the left image displays the theoretical volume simulated using a fully-resolved fluid structure interaction, which can only capture a stationary region at submicron resolution totaling a volume of 4.91 × 10−3 mL using the same compute resources. While the length scale of the fully resolved FSI model is in millimeters, the APR technique extends into meters, making it the first computational method that can track an individual cancer cell across system-scale distances while resolving submicron 3D cellular interactions.
