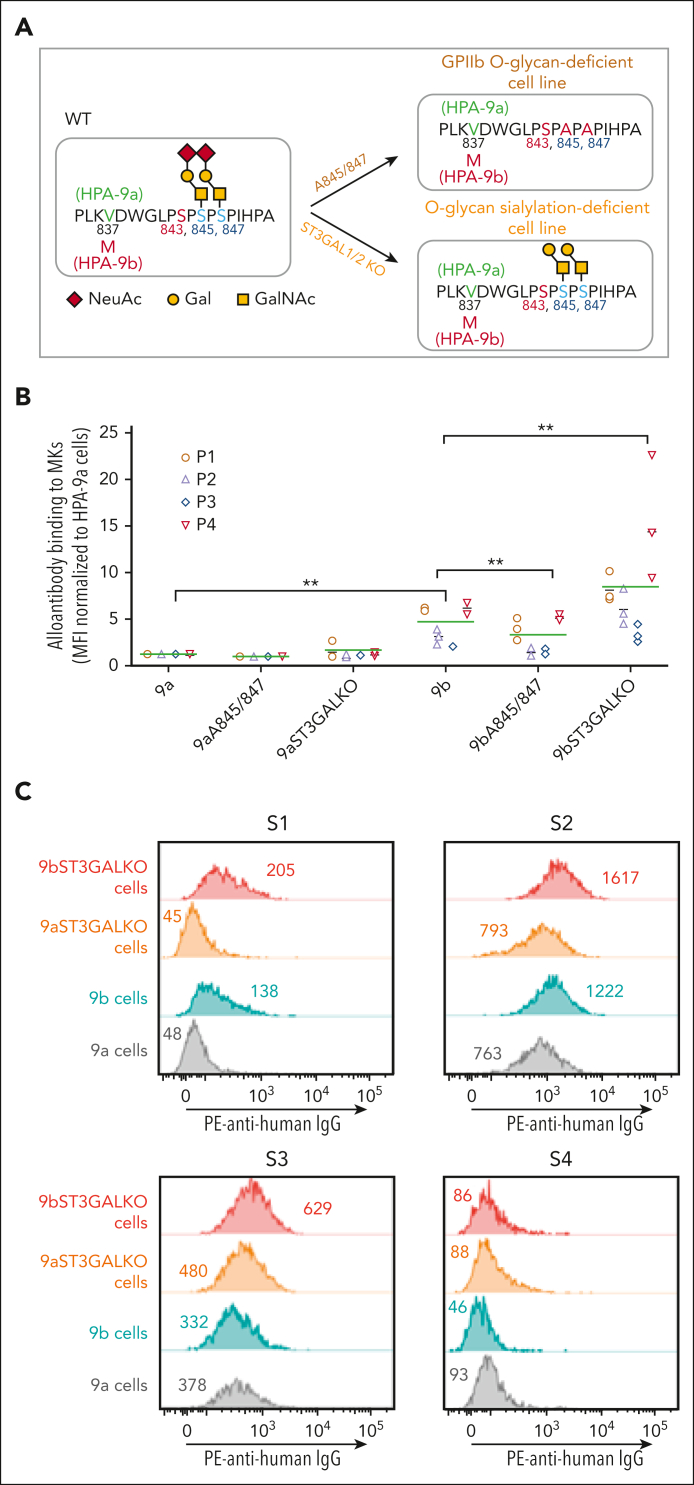
Figure 3.
Gene editing to remove terminal sialic acids or the entire O-glycans on GPIIb differentially affects anti-HPA-9b alloantibody binding. (A) Schematic of local alloantigenic peptide and O-glycan modification of GPIIb from genetically edited iPSC-derived MKs. (B) Flow cytometry analysis of the binding of 4 patient anti-HPA-9b alloantibodies to various HPA-9 allele-specific iPSC-derived MKs. Medium fluorescence intensity of each alloantibody binding to different MKs was normalized to that of WT HPA-9a cells. Green lines represent mean values of 4 patient samples in 3 independent experiments. P values are calculated from Linear mixed model fit by REML, using R package “lme4,” in which patients are random effects and cell types are fixed effects. ∗∗P < .01. (C) Screen of suspected anti-HPA-9b FNAIT maternal sera using WT or ST3GAL1/2 KO HPA-9 allele-specific iPSC-derived MKs. Eight suspected anti-HPA-9b maternal sera that were previously unconfirmable with clinical standard PABA assay were tested with whole cell flow cytometry analysis. All the suspected mothers are HPA-9a homozygous and all the fathers are HPA-9a/9b heterozygous. S1 and S2 contain anti-HPA-9b alloantibodies that are detectable by both WT and ST3GAL1/2 KO HPA-9b cells. Anti-HPA-9b alloantibody in S3 is only detectable by ST3GAL1/KO HPA-9b cells. S2 and S3 also contain anti-HPA-1a alloantibodies that cause high background binding in this assay. S4 is a representative of negative samples for the analysis. Color-coded numbers indicate median fluorescence intensity of corresponding peaks. NeuAc, N-acetylneuraminic acid; Gal, galactose; GalNAc, N-acetyl-D-galactosamine; IgG, immunoglobulin G; PE, phycoerythrin.