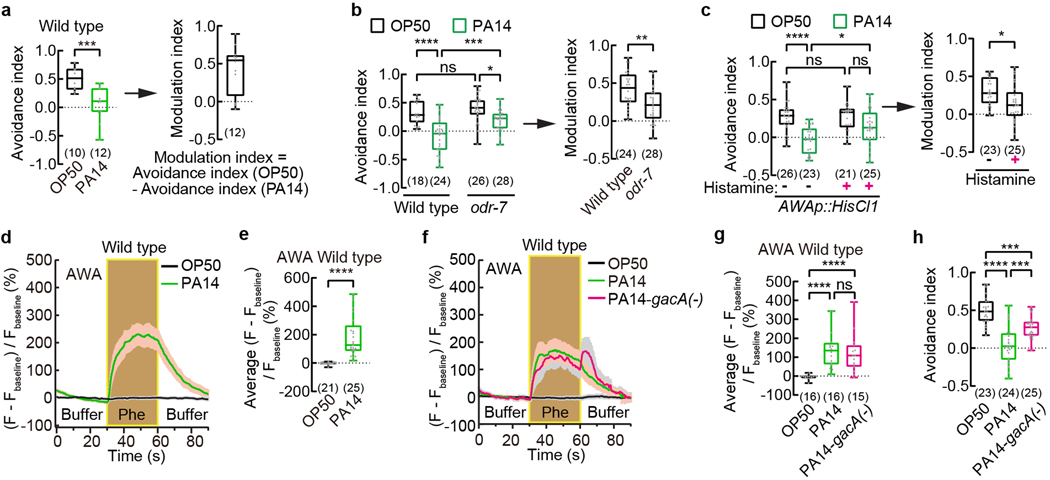
Fig. 1. Exposure to pathogenic Pseudomonas strain PA14 suppresses pheromone avoidance in hermaphrodites and induces pheromone response in AWA.
a - c, Avoidance of ascaroside pheromones when exposed to E. coli OP50 or P. aeruginosa PA14 (avoidance index) and modulation of pheromone response by PA14 (modulation index) in wild-type (a) and odr-7 mutant (b) adult C. elegans hermaphrodites, and in transgenic adult hermaphrodites expressing HisCl1 in AWA (c). Pheromones, mixture of ascr#2,3,5 (10 nM each at equilibrium). Positive avoidance index, avoidance. Positive modulation index, suppression of avoidance by PA14.
d – g, Traces of GCaMP6 signals in AWA neurons of wild-type adult hermaphrodites in response to ascaroside pheromones when exposed to different bacteria (d,f) and quantitation of average GCaMP6 signals during pheromone stimulation in the previous panel (e,g). Phe, Pheromone mixture of ascr#2,3,5 (1 μM each). Lines in traces, mean. Shades, s.e.m.. Fbaseline, average GCaMP6 signals in the first 30 seconds.
h, Avoidance of ascaroside pheromones in wild-type adult hermaphrodites exposed to E. coli OP50 or PA14 or PA14-gacA(−). Pheromones, mixture of ascr#2,3,5 (10 nM each at equilibrium). Positive avoidance index, avoidance.
Box plot, median, 1st and 3rd quartiles; whiskers, minimum and maximum. Numbers in parenthesis, number of individual assays (a-c,h) or number of individual worms (e,g). Dots, avoidance indexes or modulation indexes of individual assays (a-c,h) or average GCaMP6 signals of individual worms during pheromone stimulation (e,g). P values are derived from two-tailed unpaired t test (avoidance index in a, modulation index in b and c) or Two-way ANOVA with Tukey’s multiple comparisons test (avoidance index in b and c) or two-tailed Mann-Whitney test (e) or Kruskal-Wallis test with Dunn’s multiple comparisons test (g) or One-way ANOVA with Tukey’s multiple comparisons test (h), asterisks indicate significant difference, **** P < 0.0001, *** P < 0.001, ** P < 0.01, * P < 0.05; ns, not significant. P values are shown in Source data.