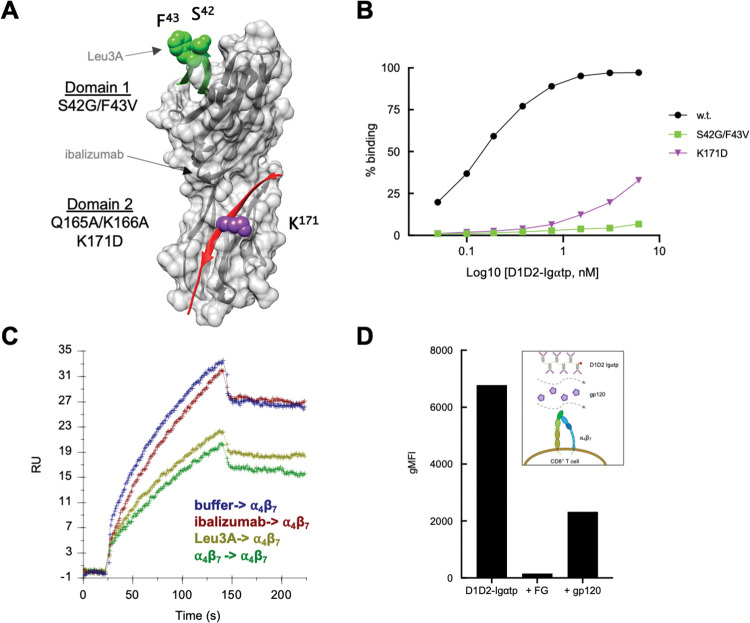
Fig 8. Mutating a gp120 contact site in CD4 D1 abrogates CD4-⍺4β7 interactions.
(A) Positions of amino acid substitutions introduced into D1D2-Ig⍺tp are indicated on a space-filling model of CD4 D1D2. Substitutions at S42/F43 in D1 (green) and K171 in D2 (purple) are shown. Approximate locations of mAb Leu3A and ibalizumab epitopes are indicated. (B) Flow-cytometric analysis of increasing amounts of D1D2-Ig⍺tp and mutant derivatives binding to ⍺4β7 on CD8+ T cells. Y-axis indicates % cells bound. (C) SPR analysis of soluble ⍺4β7 binding to a D1D2 coated surface following preincubation of the surface with ibalizumab (red), mAb Leu3A (light green), or ⍺4β7 (dark green). Y-axis indicates the mass of soluble ⍺4β7 bound at the end of the association phase. Mass of prebound inhibitors is subtracted. (D) Flow-cytometric analysis of unlabeled gp120 inhibiting PE labeled D1D2-Ig⍺tp binding to ⍺4β7 on CD8+ T cells. The integrin ⍺4 antagonist FG was employed as a control. Y-axis indicates MFI.