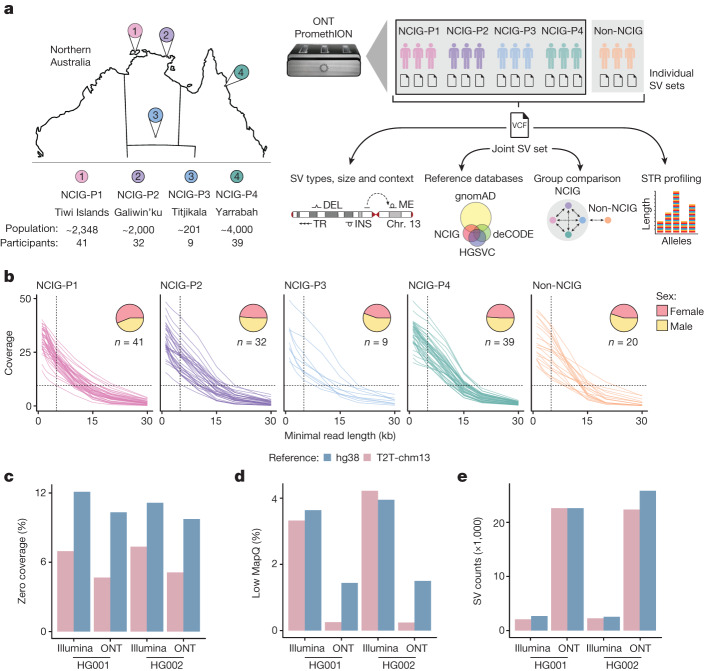
Fig. 1. Long-read sequencing in Indigenous Australian communities.
a, Study design and analysis workflow. DNA samples were collected from four Indigenous communities: Tiwi Islands (NCIG-P1), Galiwin’ku (P2), Titjikala (P3) and Yarrabah (P4), and from unrelated European individuals (non-NCIG). The map shows geographic locations, with population sizes and participant numbers underneath. ONT sequencing was performed and reads aligned to the T2T-chm13 genome. SVs were called for each individual, then joint calling was performed to generate a non-redundant set of SVs, genotyped for each individual. SVs were characterized by type, size and context and compared to existing SV datasets. SVs were compared between individuals and communities, with non-NCIG individuals as an outgroup. Short tandem repeat (STR) alleles were genotyped to assess variation. Chr, chromosome; DEL, deletions; INS, insertions; ME, mobile elements. b, Average genomic coverage as sequencing reads were filtered by a minimum read-length cut-off. Each line represents one individual. Pie charts show the proportion of male and female participants from each community. c, Percentage of genome with zero coverage for Illumina short-read and ONT long-read libraries from HG001 and HG002, aligned to either hg38 or T2T-chm13. d, Percentage of genome covered by alignments with low mapping quality (MAPQ < 5). e, Number of SVs detected.