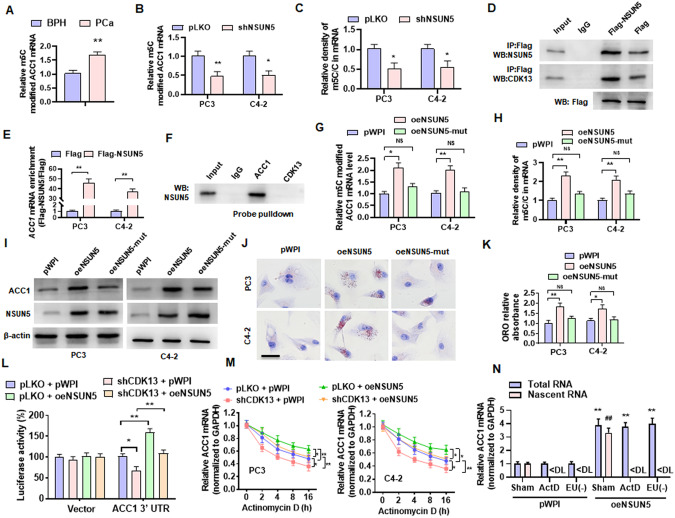
Fig. 5. NSUN5 promotes ACC1 expression and lipid deposition by regulating the m5C modification of ACC1 mRNA.
A Methylated RNA was immunoprecipitated by an m5C specific antibody (MeRIP), and then qPCR detected the level of m5C-modified ACC1 mRNA in the BPH and PCa tissues. B PC3 and C4-2 cells were transfected with shNSUN5 or control vector, and then MeRIP-qPCR examined m5C-modified ACC1 mRNA. C PC3 and C4-2 cells were ransfected as in (B) and then the UHPLC-MRM-MS/MS analysis detected the m5C methylation levels in the total mRNA. D PC3 cells were transfected with Flag-NSUN5 or a control vector (Flag), followed by CoIP to examine the interaction between CDK13 and NSUN5 and the efficacy pulled down by anti-Flag antibody. E PC3 and C4-2 cells were transfected as in (D), and RNA immunoprecipitation (RIP) with anti-Flag antibody detected the NSUN5 binding to ACC1 mRNA. F Lysates of PC3 cells were pulled down with biotinylated probe recognizing CDK13 or ACC1 mRNA, and then NSUN5 in the precipitates was detected by western blotting. G PC3 and C4-2 cells were transfected with a vector encoding wild-type or mutant forms (oeNSUN5-mut, K65A/C404, L405 del) of NSUN5, and then MeRIP-qPCR examined m5C-modified ACC1 mRNA. H PC3 and C4-2 cells were treated as in (G), and the UHPLC-MRM-MS/MS measured the m5C methylation levels in the total mRNA. I PC3 and C4-2 cells were treated as in (G), Western blot detected ACC1 and NSUN5 protein level. J, K ORO staining examined lipid accumulation in PC3 and C4-2 cells transfected with oeNSUN5 or mutant oeNSUN5. Scale bar = 20 μm. L ACC1 3’UTR was cloned into a psiCHECK™-2 Vector and then co-transfected into HEK-293T cells with shCDK13 or oeNSUN5, alone or together, and then a luciferase reporter assay detected the luciferase activity of ACC1 3ʹ-UTR luciferase reporter. M PC3 and C4-2 cells were transfected with the indicated constructs, followed by stimulation with actinomycin D for 0, 2, 4, 8 and 16 h. The ACC1 mRNA was determined by RT-qPCR. N RT-qPCR detected total and nascent ACC1 mRNA in oeNSUN5-overexpressed PC3 cells treated with or without actinomycin D or EU. ActD: actinomycin D; EU(-): without EU treatment. All data are expressed as the mean ± SEM of 3 independent experiments. *P < 0.05, #P < 0.05, ##P < 0.01 and **P < 0.01 vs. their corresponding controls.