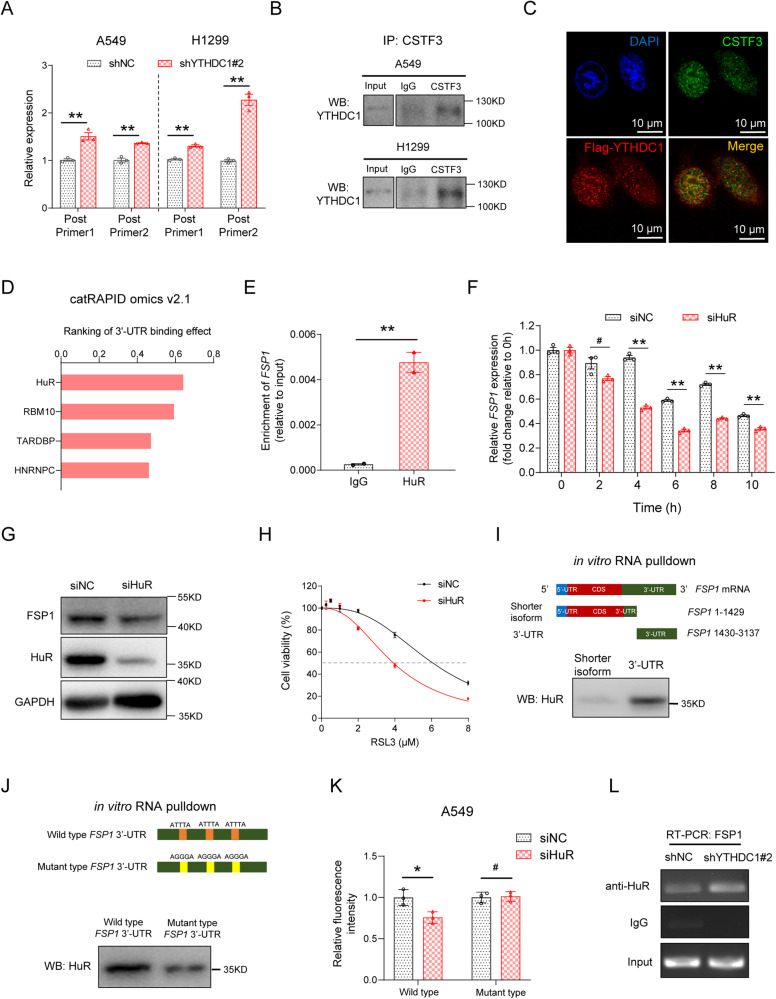
Fig. 5. YTHDC1 knockdown regulates the alternative polyadenylation and increases the long isoform of FSP1 mRNA, which can be bound and stabilized by HuR protein.
A The longer isoform of FSP1 mRNA is detected using two primers targeting 3′-UTR of FSP1 mRNA in A549 and H1299 cells. The targeting sites of the primers are shown in Fig. 4D. B The binding effects of YTHDC1 and CSTF3 are detected by Co-IP assays in A549 and H1299 cells. C The co-localization of Flag-tagged YTHDC1 and CSTF3 are detected using a confocal microscopy in A549 cells. D The binding effect of FSP1 mRNA 3′-UTR (FSP1 mRNA region: 1430-3137) to the HuR, HNRNPC, TARDBP and RBM10 proteins are predicted using catRAPID omics v2.1. E The binding effect of FSP1 mRNA by HuR protein is detected by RIP assay in A549 cells. F Stability of FSP1 mRNA over 10 h is measured by qRT-PCR relative to time 0 h after blocking new RNA synthesis with Actinomycin D (5 μg/mL) in A549 cells after knockdown of HuR. G A549 cells are transfected with siRNAs targeting HuR, and FSP1 protein levels are detected using WB. H RSL3 treatment is conducted to clarify the effect of HuR on the ferroptosis in A549 cells. Cell proliferation is detected by CCK-8 assays and IC50 curves are generated using Graphpad Prism 8.0. I The binding region of FSP1 mRNA by HuR protein is detected by RNA pulldown assays in A549 cells. J The binding effects of wild type and mutant 3’-UTR of FSP1 mRNA to HuR protein are detected by RNA pulldown assays in A549 cells. K The binding effects of wild type and mutant 3’-UTR of FSP1 mRNA to HuR protein are detected by Luciferase reporter assays in A549 cells. L The binding effects of FSP1 mRNA by HuR protein are detected by RIP assays in A549 cells with stable knockdown of YTHDC1. Results are evaluated using the two-tailed Student’s t-test. #P > 0.05; *P < 0.05; **P < 0.01.