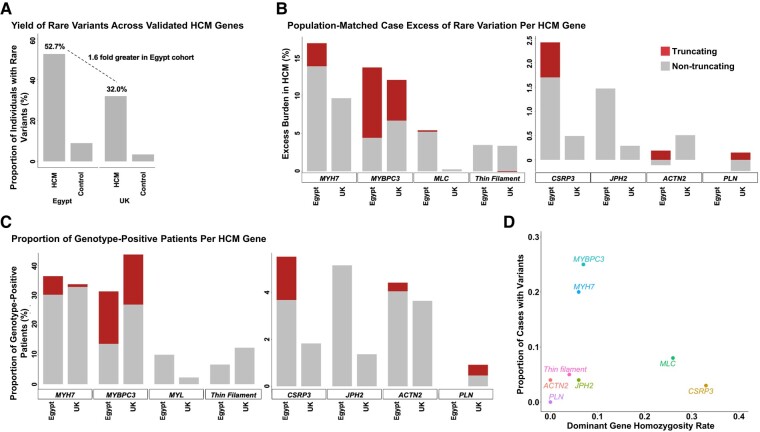
Figure 1.
Enrichment of rare variation and homozygosity in hypertrophic cardiomyopathy minor genes (MLC and CSRP3) in the Egypt hypertrophic cardiomyopathy cohort vs. UK hypertrophic cardiomyopathy. (A) The overall frequency of rare variants (gnomADpopmax FAF ≤4 × 10−5) in 13 validated hypertrophic cardiomyopathy genes was compared between Egypt and UK hypertrophic cardiomyopathy patients and controls. A 1.6-fold greater excess burden (i.e. total case frequency − total control frequency) was observed in the Egypt hypertrophic cardiomyopathy cohort, which should be accounted for with the direct gene comparisons (B). (B) Population-matched patients vs. control comparison (Egypt HCMControl excess and UK HCMControl excess) of genetic variation between Egypt and UK cohorts for each hypertrophic cardiomyopathy gene/gene class. (C) Comparison of the proportion of genotype-positive patients (i.e. those carrying rare variants) per hypertrophic cardiomyopathy gene confirms the enrichment of rare variants in the hypertrophic cardiomyopathy minor genes MLC (MYL2 and MYL3) and CSRP3 in Egyptians (as observed in B). Truncating and nontruncating variants (A, B) are shown in red and grey, respectively. Thin filament: ACTC1, TNNC1, TNNI3, TNNT2, and TPM1. (D) MLC (MYL2 and MYL3) and CSRP3 genes showed a markedly higher rate of homozygosity compared with major HCM genes (MYBPC3 and MYH7) and thin filament genes (ACTC1, TNNC1, TNNI3, TNNT2, and TPM1). HCM, hypertrophic cardiomyopathy.