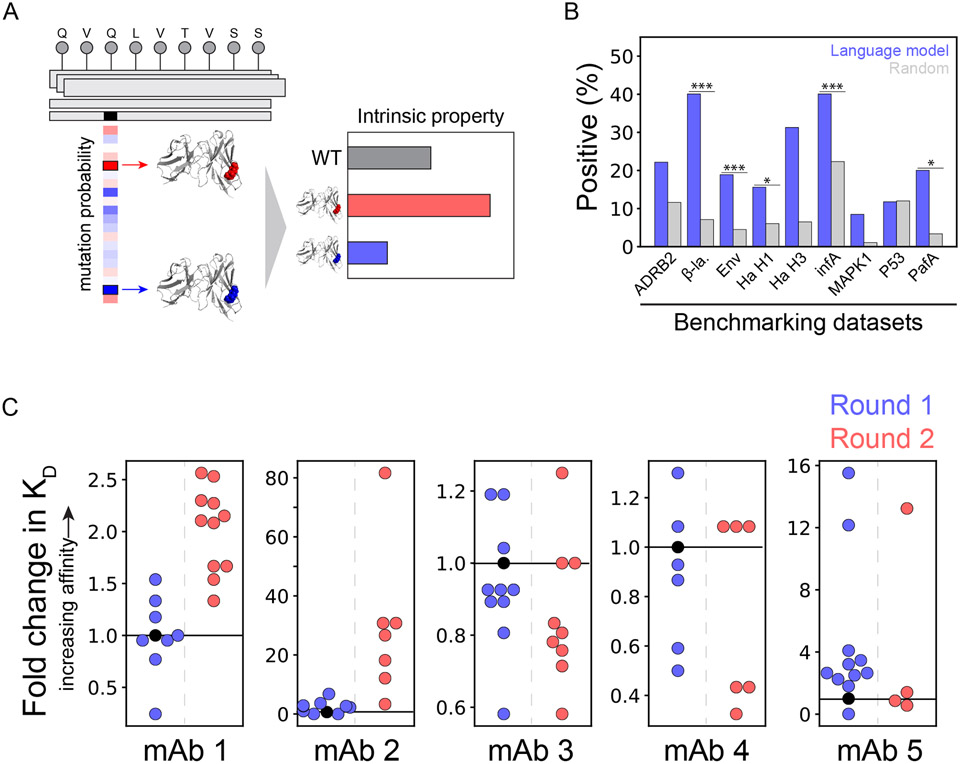
Figure 2. General protein language model identifies mutations that increase antibody affinity.
(A) Self-supervised deep transformer neural networks learn the intrinsic fitness of protein sequences and can predict mutations that increase their fitness. These intrinsic fitness predictions are hypothesized to reflect intrinsic properties of diverse proteins, including antibodies, and can be used in their optimization. (B) Benchmarking of the neural networks on nine high-throughput scanning mutagenesis datasets revealed improved or comparable predictions of single mutations that improved intrinsic properties over background predictions. (C) Mutations with higher predicted intrinsic fitness were identified and introduced to five anti-viral antibodies, resulting in higher affinities in several cases. In Round 1 of optimization, single mutations were evaluated. In Round 2 of optimization, combinations of successful mutations from Round 1 were evaluated. In (B), ADRB2 is adrenoreceptor beta 2, β-la. is β-lactamase, Env is envelope glycoprotein, Ha is hemagglutinin, infA is translation initiation factor 1, MAPK1 is mitogen-activated protein kinase 1, and PafA is phosphate-irrepressible alkaline phosphatase. Moreover, in (B), the p-values are <0.05 (*), <0.01 (**) and <0.001 (***). This figure is adapted from a previous publication.11