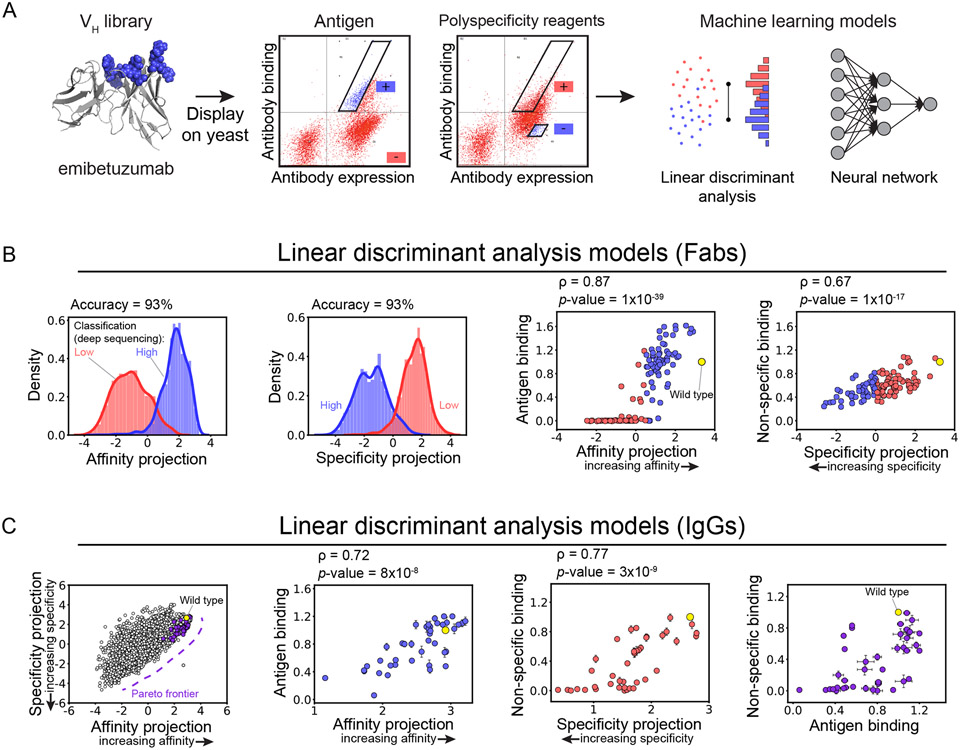
Figure 5. Linear models trained only on binary deep sequencing data predict continuous properties of a therapeutic antibody.
(A) A library of Fab variants of a therapeutic antibody (emibetuzumab) with mutations in its heavy chain CDRs was displayed on the surface of yeast and sorted for high and low levels of antigen (hepatocyte growth factor receptor, also known as c-MET) and non-specific binding. The enriched libraries were deep sequenced, and simple (linear discriminant analysis or LDA) and more complex (neural network) models were trained to classify antibody sequences for high and low levels of each property. (B) High classification accuracy was observed for the LDA models for both properties. Additionally, the LDA model projections were found to correlate with continuous measurements for both antigen and non-specific binding. (C) Co-optimized antibody mutants were identified along the Pareto frontier. These predictions were confirmed experimentally for soluble IgGs, leading to the identification of several (16) emibetuzumab variants with increased antigen and reduced non-specific binding. This figure is adapted from a previous publication.28