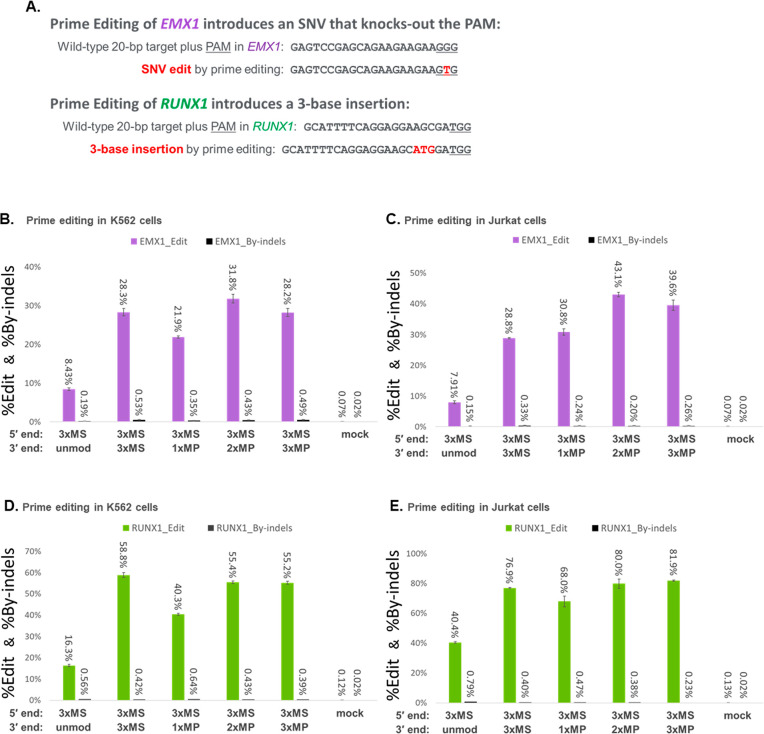
Figure 2.
Incorporation of MP or MS modifications at the 3′ end of chemically synthesized pegRNAs can improve prime editing yields. (A) Prime editing approaches were adopted to knockout the PAM in EMX1 or to introduce a three-base insertion in RUNX1. (B and D) K562 cells were cotransfected with PE2 mRNA and synthetic pegRNA modified by 3xMS at the 5′ end and various modification schemes at the 3′ end (as indicated) for editing EMX1 (B) or RUNX1 (D). (C and E) Jurkat cells were likewise transfected using the same pegRNAs for editing EMX1 (C) or RUNX1 (E). Editing yields were measured by deep sequencing of PCR amplicons of the target loci for both the desired edit (%Edit) and any contaminating indel byproducts (%By-indels). Bars represent means with standard deviation (n = 3).