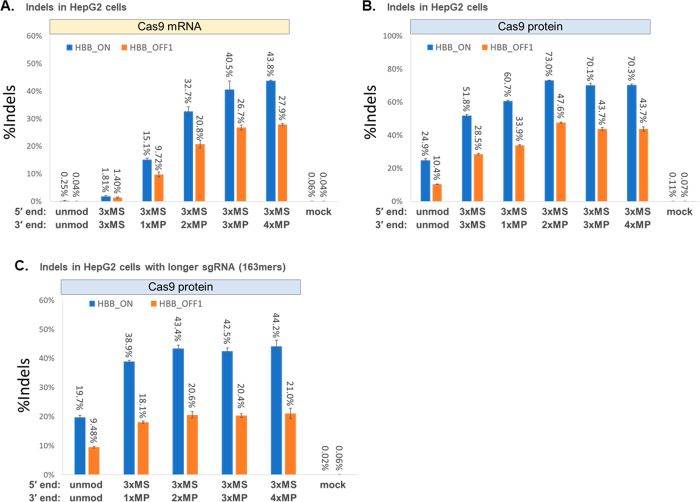
Figure 3.
Use of MP modifications at the 3′ end of chemically synthesized sgRNAs helps to maximize editing yields in the presence of serum. (A and B) To simulate a harsher biological environment for delivering CRISPR-Cas9 to cells in vivo (where bodily fluids such as serum are present), HepG2 cells were spun down from culture media without rinsing off residual bovine serum, and the cells were transfected with sgRNAs targeting the HBB locus containing different types of 3′ end modifications (as indicated) together with Cas9 mRNA (A) or were transfected with Cas9 RNP prepared by precomplexation of the sgRNA with SpCas9 protein (B). (C) Extended-length 163mer sgRNAs targeting the same HBB locus but designed for CRISPRa SAM systems were precomplexed with SpCas9 protein for targeted double-strand cleavage and indel formation by transfecting the Cas9 RNP into HepG2 cells, again without rinsing off residual serum. Indel yields were measured by deep sequencing of PCR amplicons of the target HBB locus (HBB_ON) and a known highly reactive off-target site on chromosome 9 (HBB_OFF1). Bars represent means with standard deviation (n = 3).