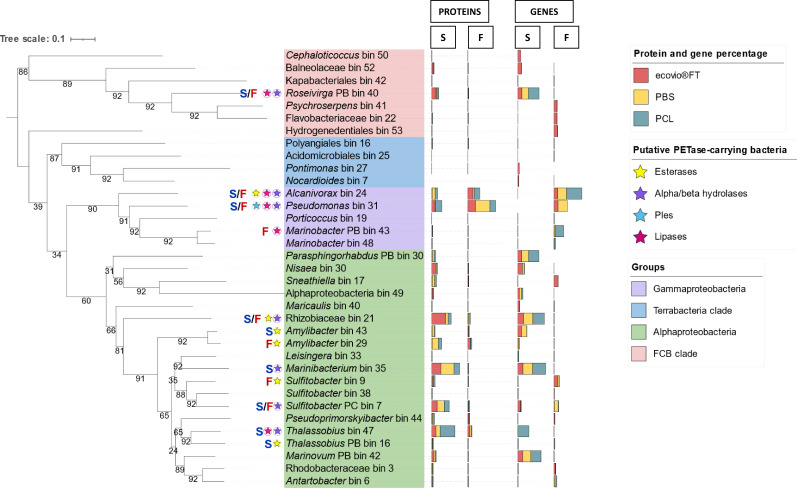
Fig 3.
Gene/protein contribution of the MAGS and their phylogenetic relationship. The 35 MAGs identified in this study and the corresponding taxa are shown. Multibars on the right side of each taxa/MAG represent percentages of either genes or proteins that were significantly (≥0.05) found in the film-attached fraction (F) or in the free-living fraction (S) in ecovio FT (light red), PBS (light yellow), and PCL (dark cyan) cultures. A star on the left side of a taxa/bin, indicates the presence of an α/β hydrolase (purple), esterase (yellow), lipase (magenta), or Ples (light blue) found in a bin and that was significantly found in the film-attached fraction (shown as “F” in red), free-living fraction (shown as “S” in blue) or both fractions (shown as “S/F” in blue and red, respectively). MAGs are additionally clustered and highlighted by the following groups: Gammaproteobacteria (soft purple), Alphaproteobacteria (soft blue), Terrabacteria (soft blue) clade, and FCB clade (soft pink). The numbers on the branches represent bootstrap values. The phylogenetic tree was created by using the UBCG pipeline and default parameters (https://www.ezbiocloud.net/tools/ubcg) and rooted with a midpoint root.