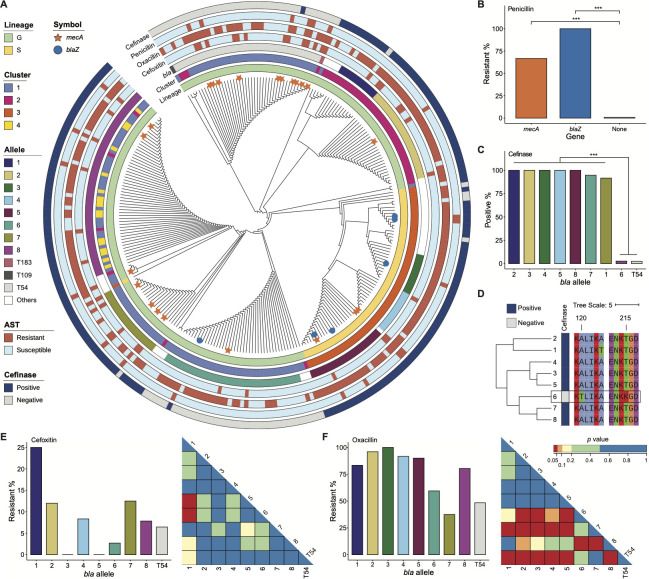
Fig 3.
Associations between gene alleles encoding β-lactamase identified in S. saprophyticus and their AMR against β-lactam antibiotics. (A) Hierarchical tree of bla sequences from all S. saprophyticus isolates, according to amino acid sequence identity. Alleles 1–8 indicate bla gene alleles with at least one amino acid substitution that each is present in at least 10 isolates, otherwise are labeled as “others.” Alleles T183, T109, and T54 indicate truncated bla based on their putative peptide length. S. saprophyticus lineages, clusters, resistance phenotypes, and β-lactamase activity are indicated by color strips. The presence of mecA and blaZ genes is symbolized by the orange star and blue circle, respectively, at the tip of the branch. Tree branch length is ignored. (B) Percentage of isolates resistant to penicillin out of the total number of S. saprophyticus carrying mecA and blaZ, compared to isolates with none of them. (C) Percentage of isolates exhibiting β-lactamase activity, detected by Cefinase assay, out of the total number of S. saprophyticus carrying specific bla gene alleles. (D) Hierarchical tree of bla gene alleles 1–8 with the alignment of amino acid sequences. β-lactamase activity is indicated by color strips. The black box highlights the unique mutations in allele 6 with the potential to influence β-lactamase activity. The scale bar represents the average number of amino acid substitutions. See Fig. S3C for the whole-length alignment. (E and F) Left panel: percentage of isolates resistant to cefoxitin and oxacillin out of the total number of mecA− S. saprophyticus carrying specific bla gene alleles. Right panel: associated P-value based on the χ2 test compared resistant rates between different alleles. The P-value is color-coded, and red indicates significant differences. The χ2 test is used with a significance threshold of 0.05 in B–C and E–F. *** P < 0.001.