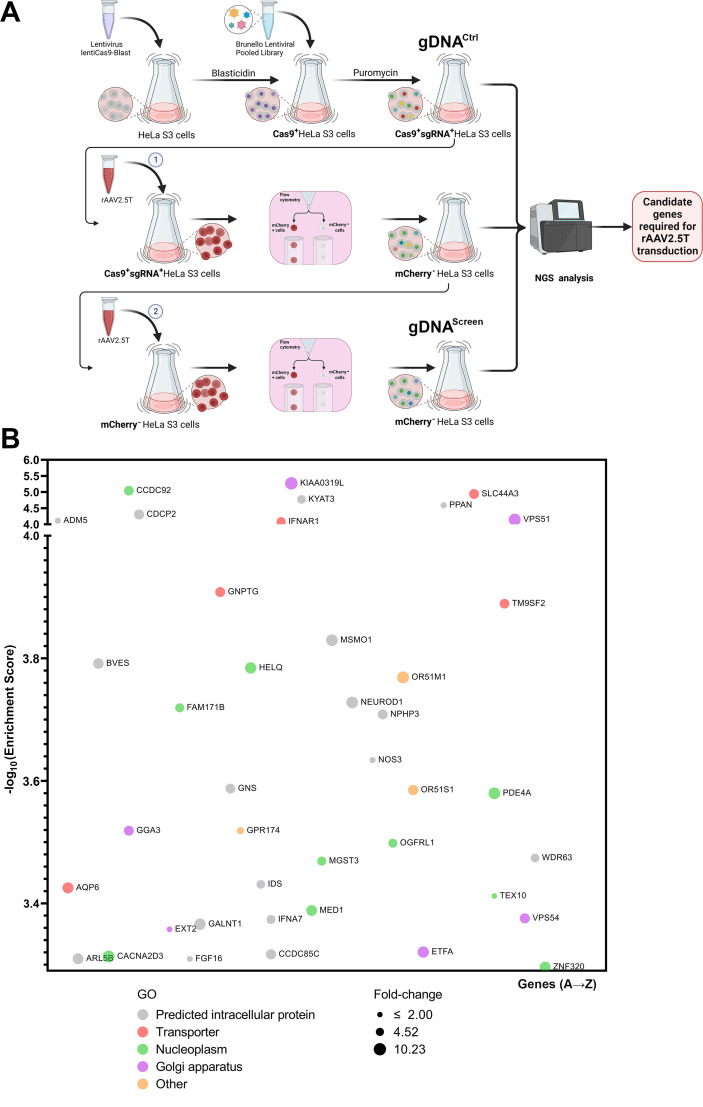
Fig 1.
Genome-wide guide RNA (gRNA) library screening of essential factors of rAAV2.5T transduction in HeLa S3 cells. (A) A diagram of the genome-wide gRNA library screening. HeLa S3 suspension cells were transduced with a spCas9-expressing lentivirus. Blasticidin-resistant spCas9-expressing cells were transduced with the Brunello lentiCRISPR gRNA lentiviral library. The library-selected HeLa S3 cells were expanded to ~200 million, of which ~100 million were extracted for genomic DNA (gDNA) as the non-selected control gDNA (gDNACtrl), and another 100 million were transduced with mCherry-expressing rAAV2.5T. At 3 days post-transduction, ~1 million mCherry-negative cells were sorted and expanded. Approximately 100 million cells were transduced with rAAV2.5T, followed by sorting of the top 1% mCherry-negative cells. The sorted mCherry-negative cells were expanded to ~100 million for extraction of gDNAScreen. The gDNA samples were subjected to NGS analysis and bioinformatics analysis. Created with BioRender. (B) Hits of host genes from the second round of screening of mCherry-negative cells. This panel shows the entry screening hits from the second-round selection of mCherry-negative cells transduced with rAAV2.5T. The x-axis shows genes within the target of the Brunello library that are grouped by gene ontology analysis. Each circle represents a gene with the size corresponding to the fold change of sgRNA reads by comparison of gDNAScreen to gDNACtrl. The y-axis shows the enrichment score (−log10) of each gene based on MAGeCK analysis of the sgRNA reads in gDNAScreen vs gDNACtrl. Only genes that have an enrichment score of >3.25 are shown. The bubble colors indicate different gene ontology classifications as shown.