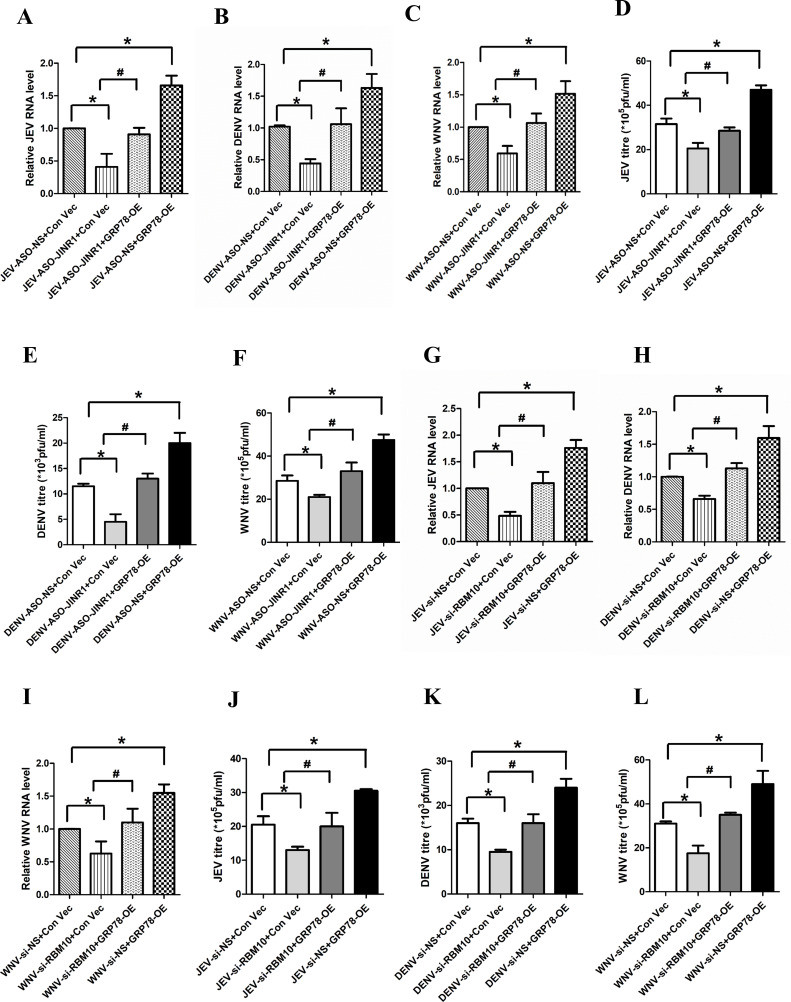
Fig 8.
GRP78 overexpression prevents the decrease in flavivirus infection due to JINR1/RBM10 knockdown. (A) GRP78 overexpression prevents the reduction in JEV RNA levels due to JINR1 depletion in SH-SY5Y cells. JEV-infected SH-SY5Y cells were co-transfected with either ASO-NS and Con-Vec or ASO-JINR1 and Con-Vec or ASO-JINR1 and GRP78-OE or ASO-NS and GRP78 OE, and viral replication was determined by quantifying the intracellular levels of JEV RNA at 48 hpi. (B) GRP78 overexpression prevents the reduction in DENV RNA levels due to JINR1 depletion in SH-SY5Y cells. DENV-infected SH-SY5Y cells were co-transfected with either ASO-NS and Con-Vec or ASO-JINR1 and Con-Vec or ASO-JINR1 and GRP78-OE or ASO-NS and GRP78 OE, and viral replication was determined by quantifying the intracellular levels of DENV RNA at 48 hpi. (C) GRP78 overexpression prevents the reduction in WNV RNA levels due to JINR1 depletion in SH-SY5Y cells. WNV-infected SH-SY5Y cells were co-transfected with either ASO-NS and Con-Vec or ASO-JINR1 and Con-Vec or ASO-JINR1 and GRP78-OE or ASO-NS and GRP78 OE, and viral replication was determined by quantifying the intracellular levels of WNV RNA at 48 hpi. (D) GRP78 overexpression prevents the reduction in JEV titer due to JINR1 depletion in SH-SY5Y cells. Quantification of viral titer from JEV-infected SH-SY5Y cells co-transfected with either ASO-NS and Con-Vec or ASO-JINR1 and Con-Vec or ASO-JINR1 and GRP78-OE or ASO-NS and GRP78 OE is shown in Fig. S23A. (E) GRP78 overexpression prevents the reduction in DENV titer due to JINR1 depletion in SH-SY5Y cells. Quantification of viral titer from DENV-infected SH-SY5Y cells co-transfected with either ASO-NS and Con-Vec or ASO-JINR1 and Con-Vec or ASO-JINR1 and GRP78-OE or ASO-NS and GRP78 OE is shown in Fig. S23B. (F) GRP78 overexpression prevents the reduction in WNV titer due to JINR1 depletion in SH-SY5Y cells. Quantification of viral titer from WNV-infected SH-SY5Y cells co-transfected with either ASO-NS and Con-Vec or ASO-JINR1 and Con-Vec or ASO-JINR1 and GRP78-OE or ASO-NS and GRP78 OE is shown in Fig. S23C. (G) GRP78 overexpression prevents the reduction in JEV RNA levels due to RBM10 depletion in SH-SY5Y cells. JEV-infected SH-SY5Y cells were co-transfected with either si-NS and Con-Vec or si-RBM10 and Con-Vec or si-RBM10 and GRP78-OE or si-NS and GRP78 OE, and viral replication was determined by quantifying the intracellular levels of JEV RNA at 36 hpi. (H) GRP78 overexpression prevents the reduction in DENV RNA levels due to RBM10 depletion in SH-SY5Y cells. DENV-infected SH-SY5Y cells were co-transfected with either si-NS and Con-Vec or si-RBM10 and Con-Vec or si-RBM10 and GRP78-OE or si-NS and GRP78 OE, and viral replication was determined by quantifying the intracellular levels of DENV RNA at 36 hpi. (I) GRP78 overexpression prevents reduction in WNV RNA levels due to RBM10 depletion in SH-SY5Y cells. WNV-infected SH-SY5Y cells were co-transfected with either si-NS and Con-Vec or si-RBM10 and Con-Vec or si-RBM10 and GRP78-OE or si-NS and GRP78 OE, and viral replication was determined by quantifying the intracellular levels of WNV RNA at 36 hpi. (J) GRP78 overexpression prevents the reduction in JEV titer due to RBM10 depletion in SH-SY5Y cells. Quantification of viral titer from JEV-infected SH-SY5Y cells co-transfected with either si-NS and Con-Vec or si-RBM10 and Con-Vec or si-RBM10 and GRP78-OE or si-NS and GRP78 OE is shown in Fig. S24A. (K) GRP78 overexpression prevents the reduction in DENV titer due to RBM10 depletion in SH-SY5Y cells. Quantification of viral titer from DENV-infected SH-SY5Y cells co-transfected with either si-NS and Con-Vec or si-RBM10 and Con-Vec or si-RBM10 and GRP78-OE or si-NS and GRP78 OE is shown in Fig. S24B. (L) GRP78 overexpression prevents the reduction in WNV titer due to RBM10 depletion in SH-SY5Y cells. Quantification of viral titer from WNV-infected SH-SY5Y cells co-transfected with either si-NS and Con-Vec or si-RBM10 and Con-Vec or si-RBM10 and GRP78-OE or si-NS and GRP78 OE is shown in Fig. S24C. Error bars represent the mean ± SEM from three independent experiments. Statistical comparison was made using the Student’s t-test. (A–C and G–I) RNA samples were analyzed by qRT-PCR. (A and D) *Significant change compared to JEV-ASO-NS + Con-Vec. #Significant change compared to JEV-ASO-JINR1 + Con-Vec. (B and E) *Significant change compared to DENV-ASO-NS + Con-Vec. #Significant change compared to DENV-ASO-JINR1 + Con-Vec. (C and F) *Significant change compared to WNV-ASO-NS + Con-Vec. #Significant change compared to WNV-ASO-JINR1 + Con-Vec. (G and J) *Significant change compared to JEV-si-NS + Con-Vec. #Significant change compared to JEV-si-RBM10 + Con-Vec. (H and K) *Significant change compared to DENV-si-NS + Con-Vec. #Significant change compared to DENV-si-RBM10 + Con-Vec. (I and L) *Significant change compared to WNV-si-NS + Con-Vec. #Significant change compared to WNV-si-RBM10 + Con-Vec.