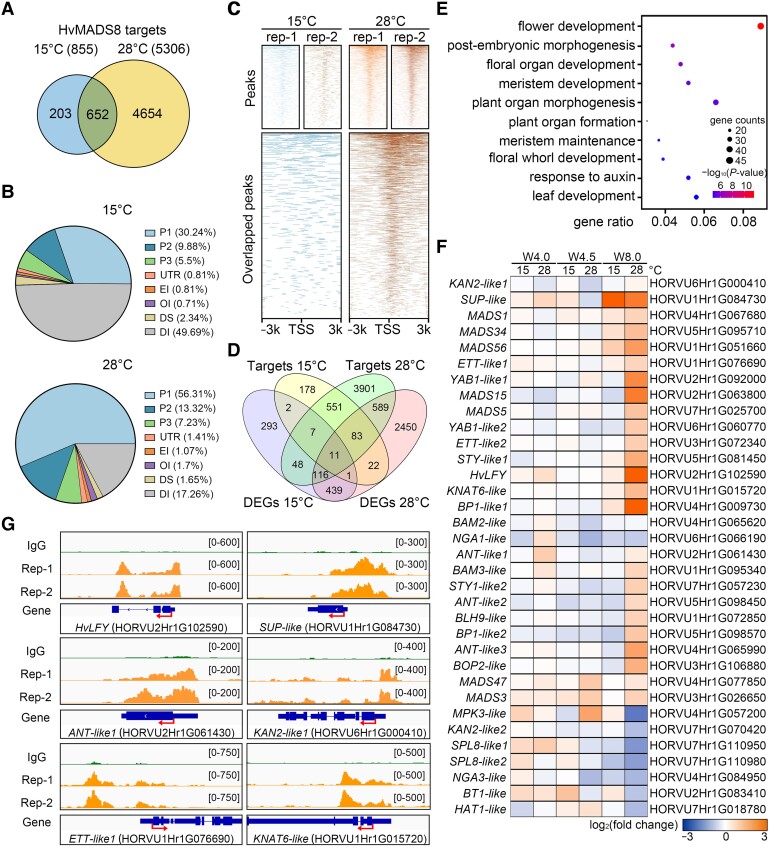
Figure 5.
Changes in HvMADS8 binding activity at high temperatures contribute to transcriptome reprogramming for floral development. A) Venn diagram showing the numbers and overlap of target genes bound by HvMADS8 at 15 and 28 °C. B) Distribution of HvMADS8-bound regions in different barley genomic regions. P1, promoters (<1 kb upstream of ATG); P2, promoters (1 to 2 kb upstream of ATG); P3, promoters (2 to 3 kb upstream of ATG); UTR, 3′ untranslated region; EI, exons and first intron; OI, other introns; DS, downstream; DI, distal intergenic. C) Density heatmaps showing CUT&Tag peaks upstream and downstream of the TSS at 15 and 28 °C. D) Venn diagram showing the overlap of HvMADS8-regulated DEGs and binding targets at 15 and 28 °C. E) GO term enrichment analysis of 799 genes common to DEGs and CUT&Tag targets at 28 °C. F) Heatmap representation of the expression of HvMADS8-bound DEGs involved in pistil and FM development. G) Chromatin-binding profiles of HvMADS8 in the promoters of 6 selected pistil regulation genes. CUT&Tag reads of 2 GFP biological repeats (Rep-1 and Rep-2; orange) and the IgG negative control (green) were visualized using the Integrative Genomics Viewer. Arrows in the gene model indicate transcription start sites, the larger boxes indicate exons, the smaller boxes illustrate untranslated regions combined from all alternative transcripts, and the lines indicate introns.