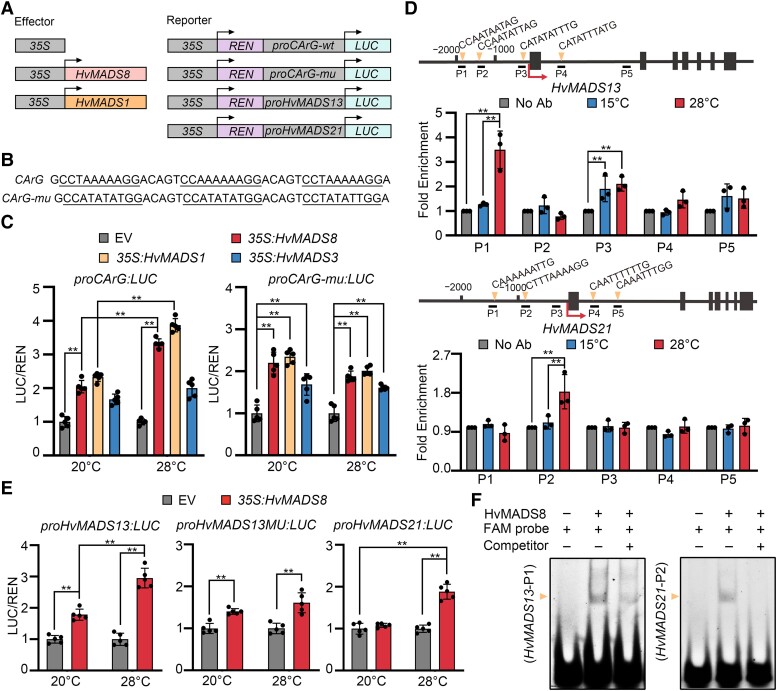
Figure 6.
HvMADS8 regulates D-class genes at high temperatures. A) Diagrams of effector and reporter constructs for dual-LUC assays. The HvMADS8 and HvMADS1 effector genes were under the control of the 35S promoter. An empty 35S effector construct was used as a negative control. LUC, firefly luciferase; REN, Renilla luciferase. B) Artificial CArG-boxes with A-tracts (underlined, CArG, WT) or non-A-tracts (underlined, CArG-mu, mutant) used to drive LUC expression. C) Normalized luciferase (LUC/REN) activity derived from the artificial CArG-box promoters in the presence of HvMADS8, HvMADS1, HvMADS3, or empty vector (EV, negative control) at 20 and 28 °C. Values are means ± Sd; n = 5 biological replicates. Asterisks indicate significant differences (2-way ANOVA test; **P < 0.01). D) In vivo binding of HvMADS13 and HvMADS21 CArG-boxes by HvMADS8 at 15 and 28 °C, as determined by ChIP-qPCR. Upper, HvMADS13 or HvMADS21 genomic regions, showing locations of 5 DNA fragments (P1 to P5) for ChIP-qPCR. Boxes indicate exons; lines indicate promoter or intron regions; arrows indicate transcription start sites. Lower, fold enrichment of 5 DNA fragments at 15 or 28 °C relative to no-antibody (Ab) control. Values are means ± Sd; n = 3 biological replicates. Asterisks indicate significant differences (2-way ANOVA test; **P < 0.01). E) Normalized luciferase activity driven by the HvMADS13 and HvMADS21 promoters in the presence of HvMADS8 or EV (negative control) at 20 and 28 °C. The CArG-box in P1 binding site of the HvMADS13 promoter is mutated to ATGTGAATAG in the proHvMADS13MU:LUC construct. Values are means ± Sd; n = 5 biological replicates. Asterisks indicate significant differences (2-way ANOVA test; **P < 0.01). F) EMSAs to detect interactions between HvMADS8 and the P1 (CArG-box motif of the HvMADS13 promoter) and P2 (CArG-box motif of the HvMADS21 promoter) probes.