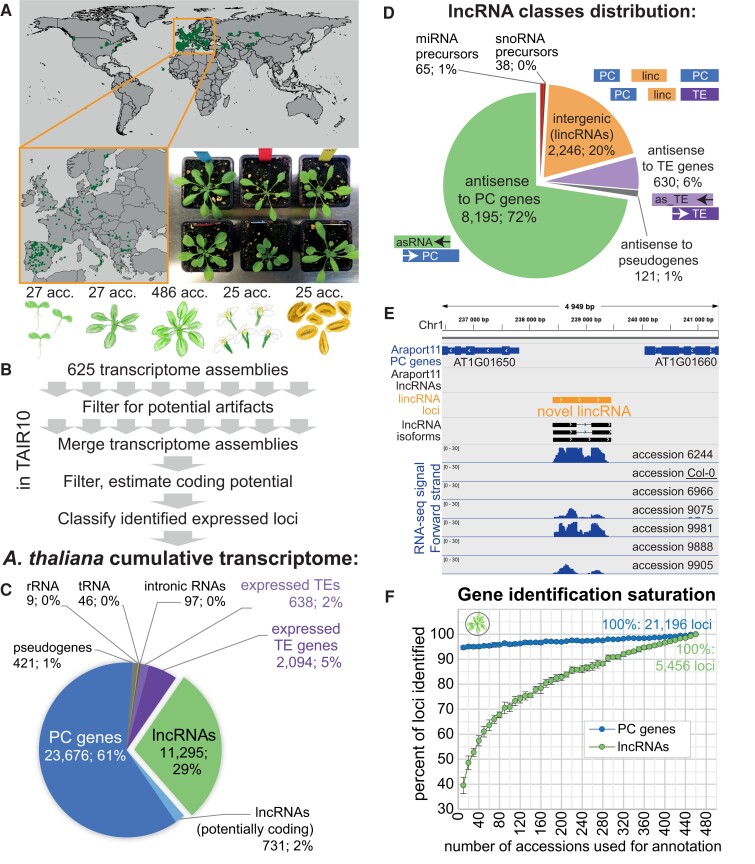
Figure 1.
Mapping lncRNA transcription in hundreds of accessions and several tissues reveals thousands of previously unannotated lncRNAs. A) Origins of the Arabidopsis accessions used for transcriptome annotation and an example photograph of 6 different accessions grown in the growth chamber. B) Overview of the pipeline used for cumulative transcriptome annotation. Tissues from left to right: 7-d-old seedlings, 9-leaf rosettes, 14-leaf rosettes, flowers, and pollen. C) Distribution of the types of loci in the cumulative annotation. D) Distribution of lncRNA positional classes in the genome. E) An example of a previously unannotated intergenic lncRNA on chromosome 1. Expression in 7 different Arabidopsis accessions is shown. F) Number of lncRNA and PC loci identified as a function of the number of accessions used, relative to the number identified using 460 accessions. Random subsampling of accessions was performed in 8 replicates, and the error bars indicate the Sd across replicates.