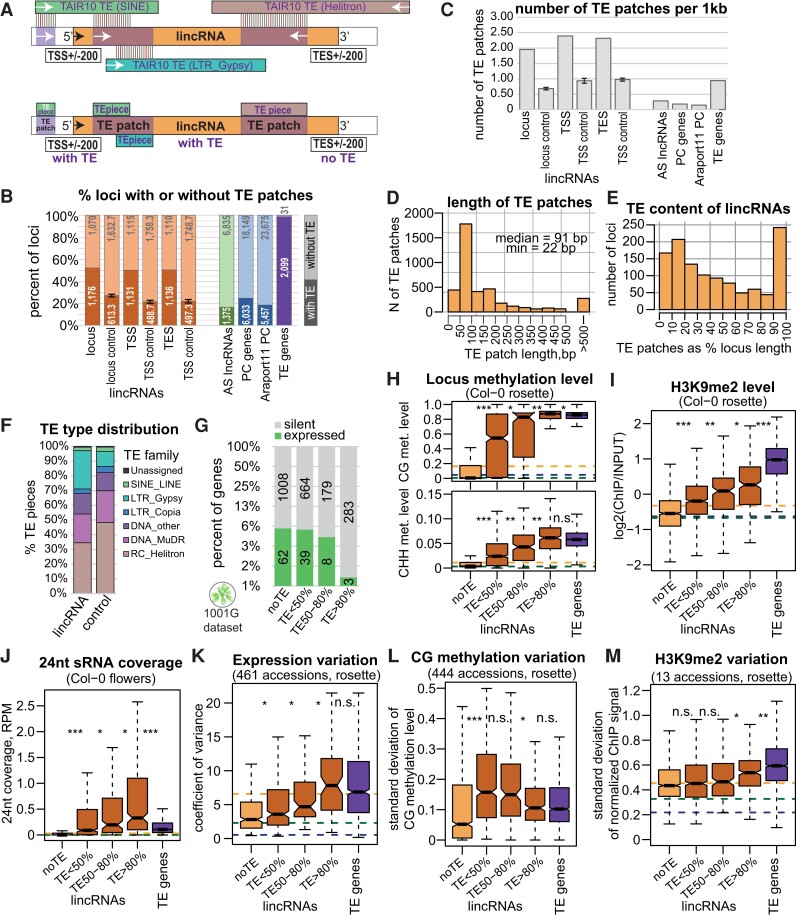
Figure 5.
Many lincRNAs contain pieces of TEs that affect their silencing and expression variation. A) Outline of TE content analysis. Top: TAIR10-annotated TEs were compared with the sequences of lincRNAs (and other loci) by BLAST. Bottom: the mapped pieces of different TEs overlapping in the same direction were merged into “TE patches.” The upstream and downstream “borders” of genes were analyzed in the same way. B) Proportion of loci containing a TE piece. The intergenic controls for lincRNAs, lincRNA TSS ± 200 bp, and TES ± 200 bp were obtained by shuffling the corresponding loci within intergenic regions (lincRNAs excluded) 3 times and averaging the results. The error bars on controls represent the Sd between the 3 shuffling replicates. C) Number of TE patches per 1 kb. D) Distribution of the length of TE patches (any relative direction) within lincRNA loci. E) TE content distribution among lincRNAs. TE patches were considered in any relative direction. The loci with large TE content are those where the TE patches map AS to the lincRNA locus. F) Proportion of TE pieces of different TE families within different types of loci. G) Proportion of expressed lincRNAs as a function of their TE content. The y axis is displayed in log2 scale. H) to J) Levels of methylation (H), H3K9me2 (I), and 24-nt sRNAs (J) for lincRNA loci as a function of their TE content, with TE genes for comparison. CG and CHH methylation data displayed are from Col-0 rosettes (Kawakatsu et al. 2016). K) to M) Expression variability between 461 accessions (Kawakatsu et al. 2016) (K), Sd of CG methylation levels across 444 accessions (Kawakatsu et al. 2016) (L), and Sd of quantile- and input-normalized H3K9me2 levels in rosettes across 13 accessions (M) of lincRNA loci as a function of their TE content, with TE genes for comparison. P-values were calculated using Mann–Whitney tests: ***P < 10–10, **P < 10–5, *P < 0.01, n.s. P > 0.01. Outliers in the boxplots are not shown.